bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2614_orf2
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
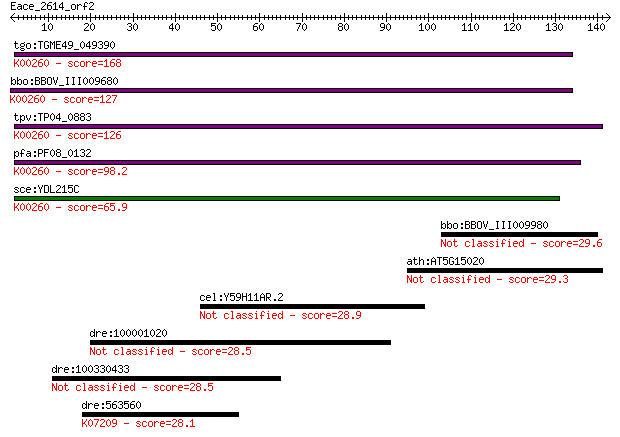
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 168 6e-42
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 127 1e-29
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 126 3e-29
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 98.2 7e-21
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 65.9 4e-11
bbo:BBOV_III009980 17.m10623; hypothetical protein 29.6 3.7
ath:AT5G15020 SNL2; SNL2 (SIN3-LIKE 2) 29.3 4.1
cel:Y59H11AR.2 hypothetical protein 28.9 5.6
dre:100001020 NLR family, pyrin domain containing 6-like 28.5 6.7
dre:100330433 thioredoxin domain containing 11-like 28.5 7.6
dre:563560 ikbkb; inhibitor of kappa light polypeptide gene en... 28.1 8.3
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 168 bits (425), Expect = 6e-42, Method: Composition-based stats.
Identities = 79/132 (59%), Positives = 99/132 (75%), Gaps = 0/132 (0%)
Query 2 FYKEYVKAILERIRENANLEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCLW 61
FY+ YV+ ++ERIRENA LEF A+W E+ RTG CDLTD+LS KI+ L DI KS+ LW
Sbjct 969 FYQTYVRQVMERIRENARLEFNALWEESLRTGKPRCDLTDVLSAKILRLNQDIHKSDSLW 1028
Query 62 ADEGLVNYVLSSAVPDVLVPQLLSVEVFRKRVPEAYQQAIFTSFLSSRFFYGQPFTANVS 121
D+ LV+ VL A+PDVLVP L+S+E R+RVPE+Y QAIF FL+SRF+Y Q FT N+S
Sbjct 1029 QDDELVSCVLLKALPDVLVPGLMSIETLRQRVPESYLQAIFQCFLASRFYYSQQFTDNLS 1088
Query 122 AFAFIAYLEELK 133
AFAF Y+ LK
Sbjct 1089 AFAFFDYVRHLK 1100
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 127 bits (319), Expect = 1e-29, Method: Composition-based stats.
Identities = 64/133 (48%), Positives = 86/133 (64%), Gaps = 1/133 (0%)
Query 1 RFYKEYVKAILERIRENANLEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCL 60
+F K+Y+ IL+ I++NA EF A+W E R G+ CDLTD+LS KII LK DI SN L
Sbjct 894 QFRKDYINEILDIIKKNARREFHALWNEGLRIGMPRCDLTDVLSTKIIRLKKDIIDSNSL 953
Query 61 WADEGLVNYVLSSAVPDVLVPQLLSVEVFRKRVPEAYQQAIFTSFLSSRFFYGQPFTANV 120
W D LV VLS A+P L +L+ +E R+P+ Y +++F S L+S F+Y Q FT +
Sbjct 954 WEDTVLVRAVLSKAIPQSL-QKLVPIEDIMVRLPDRYMRSMFASHLASTFYYSQQFTDDT 1012
Query 121 SAFAFIAYLEELK 133
S FAF YL+ LK
Sbjct 1013 SVFAFYEYLKALK 1025
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 126 bits (316), Expect = 3e-29, Method: Composition-based stats.
Identities = 65/139 (46%), Positives = 93/139 (66%), Gaps = 1/139 (0%)
Query 2 FYKEYVKAILERIRENANLEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCLW 61
F K+YV I+E IR+NA LEFEA+W E RTG+ CDLTD+LS KI LK+ I SN L+
Sbjct 1019 FRKKYVNDIMEIIRKNATLEFEALWSEGLRTGIPRCDLTDVLSDKINALKSRIKSSNTLF 1078
Query 62 ADEGLVNYVLSSAVPDVLVPQLLSVEVFRKRVPEAYQQAIFTSFLSSRFFYGQPFTANVS 121
D+ LV+ VLS VP L+ +L++VE +R+P+ Y +A+F S+++S F+Y + F+ + S
Sbjct 1079 NDKELVHTVLSRCVPASLL-ELVTVEKIVERLPQIYLRALFASYIASNFYYKEKFSDDTS 1137
Query 122 AFAFIAYLEELKQKIQKQN 140
FAF Y+ LK +N
Sbjct 1138 VFAFYEYITSLKDPAGHKN 1156
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 98.2 bits (243), Expect = 7e-21, Method: Composition-based stats.
Identities = 52/134 (38%), Positives = 80/134 (59%), Gaps = 1/134 (0%)
Query 2 FYKEYVKAILERIRENANLEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCLW 61
FYK YVK I ++I LEFE++W+E +RT +ILS KI ELK DI S+ L
Sbjct 1263 FYKAYVKEIQKKITHYCELEFESLWKETRRTKTPISKAINILSNKISELKKDILSSDTLC 1322
Query 62 ADEGLVNYVLSSAVPDVLVPQLLSVEVFRKRVPEAYQQAIFTSFLSSRFFYGQPFTANVS 121
D L+ VL +P L+ ++++ E +RVP Y +++F S L+S ++Y Q F ++S
Sbjct 1323 RDYKLLKKVLERVIPPTLL-KIVTFEQILERVPYVYIKSLFASSLASNYYYSQQFLNDLS 1381
Query 122 AFAFIAYLEELKQK 135
AF F Y+ +L+ +
Sbjct 1382 AFNFFEYITKLQSE 1395
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 68/132 (51%), Gaps = 4/132 (3%)
Query 2 FYKEYVKAILERIRENANLEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCLW 61
YK YV + RI++NA LEF +W + G ++++ LS I +L D+ S LW
Sbjct 955 LYKSYVVEVQSRIQKNAELEFGQLWNLNQLNGTHISEISNQLSFTINKLNDDLVASQELW 1014
Query 62 -ADEGLVNY-VLSSAVPDVLVPQLLSVEVFRKRVPEAYQQAIFTSFLSSRFFYGQPFTAN 119
D L NY +L +P +L+ + + + +PE+Y + + +S+LSS F Y N
Sbjct 1015 LNDLKLRNYLLLDKIIPKILI-DVAGPQSVLENIPESYLKVLLSSYLSSTFVYQNGIDVN 1073
Query 120 VSAF-AFIAYLE 130
+ F FI L+
Sbjct 1074 IGKFLEFIGGLK 1085
> bbo:BBOV_III009980 17.m10623; hypothetical protein
Length=428
Score = 29.6 bits (65), Expect = 3.7, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 103 TSFLSSRFFYGQPFTANVSAFAFIAYLEELKQKIQKQ 139
T +LS F PFTA++S+ F+ YL + Q Q++
Sbjct 157 TLYLSHHVFNPTPFTASLSSHVFVFYLPKSGQVNQRK 193
> ath:AT5G15020 SNL2; SNL2 (SIN3-LIKE 2)
Length=1367
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 13/46 (28%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 95 EAYQQAIFTSFLSSRFFYGQPFTANVSAFAFIAYLEELKQKIQKQN 140
+ Y + + S F++ Q + N+SA A ++ +++LK+K QK++
Sbjct 627 DVYAKNHYKSLDHRSFYFKQQDSKNLSAKALVSEVKDLKEKSQKED 672
> cel:Y59H11AR.2 hypothetical protein
Length=1127
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 46 KIIELKTDISKSNCLWADEGLVNYVLSSAVPDVLVPQLLSVEVFRKRVPEAYQ 98
KI++L T ++ +C +A G V++ PD+L +L VF + PE Q
Sbjct 779 KIVDL-TKMTNKDCQFAISGSTFSVVTHEYPDLLDQLVLVCNVFARMAPEQKQ 830
> dre:100001020 NLR family, pyrin domain containing 6-like
Length=1109
Score = 28.5 bits (62), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 34/75 (45%), Gaps = 7/75 (9%)
Query 20 LEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCLWADEGLVNYVLSSA----V 75
+ A+ + +KR L+CC+LT Q L + + SNC+ + L N L + +
Sbjct 724 IRLSAVIKSSKRALLQCCNLT---VQSCESLSSALQSSNCVLRELDLSNNDLQDSGVKLL 780
Query 76 PDVLVPQLLSVEVFR 90
D L Q +E R
Sbjct 781 SDGLKIQHCKLETLR 795
> dre:100330433 thioredoxin domain containing 11-like
Length=775
Score = 28.5 bits (62), Expect = 7.6, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 27/54 (50%), Gaps = 4/54 (7%)
Query 11 LERIRENANLEFEAIWREAKRTGLRCCDLTDILSQKIIELKTDISKSNCLWADE 64
L R+RE + + + +WRE +R L T L + EL+ I + L+ ++
Sbjct 656 LHRVRERVSQQLDVLWRENRRLTLH----THTLQSQNQELQAQIHQLEALYREK 705
> dre:563560 ikbkb; inhibitor of kappa light polypeptide gene
enhancer in B-cells, kinase beta (EC:2.7.11.10); K07209 inhibitor
of nuclear factor kappa-B kinase subunit beta [EC:2.7.11.10]
Length=779
Score = 28.1 bits (61), Expect = 8.3, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Query 18 ANLEFEAIWREAKRTGLRCCDLTDI--LSQKIIELKTDI 54
A+ + ++WRE ++T L C D+ L ++++ L+TDI
Sbjct 535 ASEKLLSVWREMEQTALDCGQTEDVFKLEEEMMTLQTDI 573
Lambda K H
0.323 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40