bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2636_orf2
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
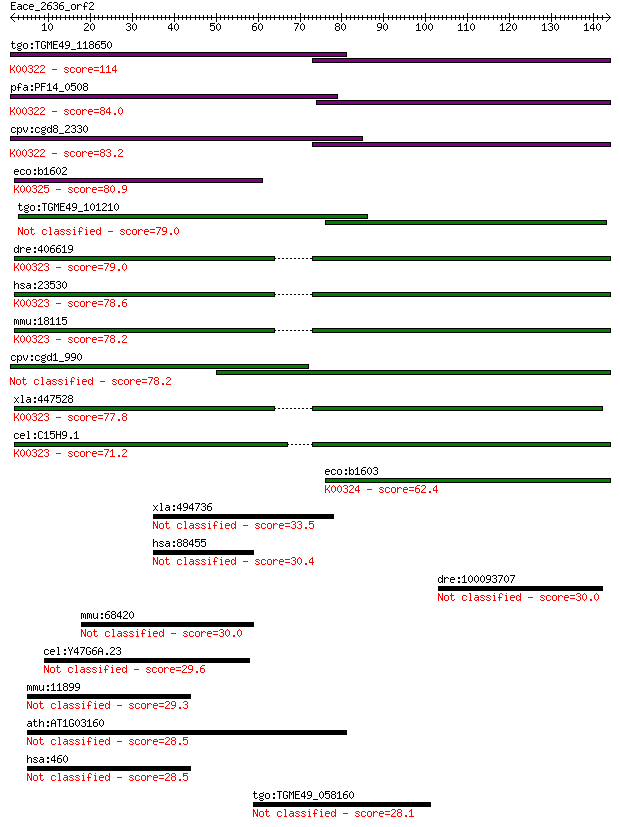
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 114 2e-25
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 84.0 1e-16
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 83.2 3e-16
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 80.9 1e-15
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 79.0 5e-15
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 79.0 5e-15
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 78.6 7e-15
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 78.2 8e-15
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 78.2 8e-15
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 77.8 1e-14
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 71.2 1e-12
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 62.4 4e-10
xla:494736 ankrd13a; ankyrin repeat domain 13A 33.5 0.25
hsa:88455 ANKRD13A, ANKRD13, NY-REN-25; ankyrin repeat domain 13A 30.4 1.8
dre:100093707 wu:fj43a03; zgc:165628 30.0 2.3
mmu:68420 Ankrd13a, 1100001D10Rik, ANKRD13, AU046136, MGC11850... 30.0 2.8
cel:Y47G6A.23 lpd-3; LiPid Depleted family member (lpd-3) 29.6 2.9
mmu:11899 Astn1, Astn, GC14, mKIAA0289; astrotactin 1 29.3
ath:AT1G03160 FZL; FZL (FZO-LIKE); GTP binding / GTPase/ thiam... 28.5 6.4
hsa:460 ASTN1, ASTN, KIAA1747; astrotactin 1 28.5
tgo:TGME49_058160 hypothetical protein 28.1 9.2
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/80 (67%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 LEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVF 60
LEPG+KI GMPVIE WKA+RV VLKRS+A GYA+I+NPLF L+NTRMLFGNAK TT+A+F
Sbjct 393 LEPGTKIYGMPVIEVWKAKRVIVLKRSLAPGYAAIDNPLFFLDNTRMLFGNAKDTTTAIF 452
Query 61 ARVNARAEQMPPSAARDDLE 80
A ++ RA + SA DLE
Sbjct 453 ACLSQRAAFVGKSAVFLDLE 472
Score = 84.7 bits (208), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 42/71 (59%), Positives = 59/71 (83%), Gaps = 0/71 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+A R+++ESCGG F+ + + EE EV GGYAREMG+AY+ AQ+++++ + + DV+ICTAA
Sbjct 695 AATREEVESCGGTFLSVELEEEGEVEGGYAREMGEAYEMAQKQMLSRVVPNVDVIICTAA 754
Query 133 IHGKPSPKLIS 143
IHGKPSPKLIS
Sbjct 755 IHGKPSPKLIS 765
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 36/78 (46%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 1 LEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVF 60
L+P SKI GMPVIE WK+++V V KR++ GY++I+NPLF+ NT +LFG+AK TT+ +
Sbjct 497 LDPSSKIYGMPVIEVWKSKQVIVFKRTLNTGYSAIDNPLFYFSNTFLLFGDAKHTTNQIL 556
Query 61 ARVNARAEQMPPSAARDD 78
+N P + D
Sbjct 557 TILNDYVNNKYPDISDQD 574
Score = 58.9 bits (141), Expect = 6e-09, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 74 AARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAAI 133
A ++++SCGG FI + E ++L + Y + Q L IK CD++IC+A+I
Sbjct 843 ATEEEVKSCGGIFIRIPTSERGDILNMSTDMNNEEYIKVQSNLFKKIIKKCDILICSASI 902
Query 134 HGKPSPKLIS 143
GK SPKL++
Sbjct 903 PGKTSPKLVT 912
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 54/84 (64%), Gaps = 0/84 (0%)
Query 1 LEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVF 60
LE SKI+GMPVIE W+A +V V KRS+ GYA+I+NPLF ++N M+FGNAK + +
Sbjct 487 LEKDSKINGMPVIEVWRASKVIVSKRSLGKGYAAIDNPLFFMKNVEMIFGNAKDSMINIL 546
Query 61 ARVNARAEQMPPSAARDDLESCGG 84
+ + + + + DD E+ G
Sbjct 547 QNIISISPEQKKANLNDDQETIDG 570
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+A R+++ES G KF+ + + E+ + GYA+ M Y +AQ +L + I+ CDVVI TA
Sbjct 785 TATREEVESLGAKFVTVDIKEDGDSGSGYAKVMSPEYLKAQSKLYSKMIRSCDVVITTAL 844
Query 133 IHGKPSPKLIS 143
I GKPSPK+I+
Sbjct 845 IPGKPSPKIIT 855
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 34/59 (57%), Positives = 44/59 (74%), Gaps = 0/59 (0%)
Query 2 EPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVF 60
+P S I+GMPV+E WKA+ V V KRSM GYA ++NPLF ENT MLFG+AK++ A+
Sbjct 401 DPKSPIAGMPVLEVWKAQNVIVFKRSMNTGYAGVQNPLFFKENTHMLFGDAKASVDAIL 459
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 51/83 (61%), Gaps = 0/83 (0%)
Query 3 PGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVFAR 62
PG I GMPV+E WKA++V VLKRSM GYA ++NPLF N+ ML G+AK++ +
Sbjct 387 PGCPIYGMPVLEVWKAKKVIVLKRSMRVGYAGVDNPLFFYPNSEMLLGDAKASLQTLLTD 446
Query 63 VNARAEQMPPSAARDDLESCGGK 85
+ + + P A+R E+ K
Sbjct 447 MQDKLDAHPIEASRLSSETQTNK 469
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 43/71 (60%), Gaps = 4/71 (5%)
Query 76 RDDLESCGGKFIGLRM---GEEAEVL-GGYAREMGDAYQRAQRELIANTIKHCDVVICTA 131
++ +ES GG+F+ + GE GGYA+ M + + R + EL A + D++I TA
Sbjct 798 KEQVESMGGEFLAMTFEGPGESGRATAGGYAKPMSEEFLRKEMELFAEQCREIDILITTA 857
Query 132 AIHGKPSPKLI 142
++ G+P+PKLI
Sbjct 858 SLPGRPAPKLI 868
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 32/62 (51%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 2 EPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVFA 61
+P S I+GMPV+E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A
Sbjct 1015 DPNSIIAGMPVLEVWKSKQVVVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDALSA 1074
Query 62 RV 63
+V
Sbjct 1075 KV 1076
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 0/71 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+AA + +S G + + + + E E GGYA+EM + A+ +L A D++I TA
Sbjct 256 AAALEQFKSLGAEPLEVDIKESGEGQGGYAKEMSKEFIEAEMKLFAKQCLDVDIIITTAL 315
Query 133 IHGKPSPKLIS 143
I G+ +P LI+
Sbjct 316 IPGRKAPVLIT 326
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 32/62 (51%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 2 EPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVFA 61
+P S I+GMPV+E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A
Sbjct 1019 DPNSIIAGMPVLEVWKSKQVIVMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQA 1078
Query 62 RV 63
+V
Sbjct 1079 KV 1080
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 0/71 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+AA + +S G + + + + E E GGYA+EM + A+ +L A K D++I TA
Sbjct 260 AAALEQFKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFAQQCKEVDILISTAL 319
Query 133 IHGKPSPKLIS 143
I GK +P L +
Sbjct 320 IPGKKAPVLFN 330
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 78.2 bits (191), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 32/62 (51%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 2 EPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVFA 61
+P S I+GMPV+E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A
Sbjct 1019 DPNSIIAGMPVLEVWKSKQVIVMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQA 1078
Query 62 RV 63
+V
Sbjct 1079 KV 1080
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 40/71 (56%), Gaps = 0/71 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+AA + +S G + + + + E E GGYA+EM + A+ +L A K D++I TA
Sbjct 260 AAALEQFKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFAQQCKEVDILISTAL 319
Query 133 IHGKPSPKLIS 143
I GK +P L S
Sbjct 320 IPGKKAPVLFS 330
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 78.2 bits (191), Expect = 8e-15, Method: Composition-based stats.
Identities = 38/71 (53%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 1 LEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVF 60
L+P SKISGMPVI WKA++V V KRS+A GYA IEN LF E TRML G+++ T V+
Sbjct 450 LDPQSKISGMPVINVWKAKQVVVSKRSLAYGYACIENELFTCERTRMLLGDSRETLQQVY 509
Query 61 ARVNARAEQMP 71
+ A +P
Sbjct 510 KILKGCAGFVP 520
Score = 36.2 bits (82), Expect = 0.040, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 48/94 (51%), Gaps = 15/94 (15%)
Query 50 GNAKSTTSAVFARVNARAEQMPPSAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAY 109
AKS + VFA +++R S ++++ ESCG +FI ++ E E L +
Sbjct 782 ATAKSMGTKVFA-MDSR------STSKEEAESCGARFI--QVPSEGESL------RKEEI 826
Query 110 QRAQRELIANTIKHCDVVICTAAIHGKPSPKLIS 143
+ QR+LI + D+VI +A G+ P LI+
Sbjct 827 LKKQRDLIEKYLCISDIVITSACKPGEECPILIT 860
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 2 EPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVFA 61
+P S I+GMPV+E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T ++ A
Sbjct 1019 DPNSIIAGMPVLEVWKSKQVIVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDSLQA 1078
Query 62 RV 63
+V
Sbjct 1079 KV 1080
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 39/69 (56%), Gaps = 0/69 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+AA + +S G + + + + E E GGYA+EM + A+ +L A + D+++ TA
Sbjct 260 AAALEQFKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFAKQCQDVDIIVTTAL 319
Query 133 IHGKPSPKL 141
I GK +P L
Sbjct 320 IPGKTAPIL 328
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 2 EPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKSTTSAVFA 61
+P S I+GMPV+ W +++V ++KR++ GYA+++NP+F ENT+ML G+AK + +
Sbjct 975 DPNSSIAGMPVLRVWNSKQVIIVKRTLGTGYAAVDNPVFFNENTQMLLGDAKKMSEKLLE 1034
Query 62 RVNAR 66
V ++
Sbjct 1035 EVKSK 1039
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 45/71 (63%), Gaps = 0/71 (0%)
Query 73 SAARDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 132
+A ++ +ES G +F+ + + E+ E GGYA+EM + A+ +L A+ K D++I TA
Sbjct 226 AAVKEHVESLGAQFLTVNVKEDGEGGGGYAKEMSKEFIDAEMKLFADQCKDVDIIITTAL 285
Query 133 IHGKPSPKLIS 143
I GK +P LI+
Sbjct 286 IPGKKAPILIT 296
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 44/68 (64%), Gaps = 0/68 (0%)
Query 76 RDDLESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAAIHG 135
++ ++S G +F+ L EEA GYA+ M DA+ +A+ EL A K D+++ TA I G
Sbjct 201 KEQVQSMGAEFLELDFKEEAGSGDGYAKVMSDAFIKAEMELFAAQAKEVDIIVTTALIPG 260
Query 136 KPSPKLIS 143
KP+PKLI+
Sbjct 261 KPAPKLIT 268
> xla:494736 ankrd13a; ankyrin repeat domain 13A
Length=593
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 9/43 (20%)
Query 35 IENPLFHLENTRMLFGNAKSTTSAVFARVNARAEQMPPSAARD 77
IE PLFH+ N R+ FGN + + RAE P S R+
Sbjct 411 IEIPLFHVLNARITFGNVNTCS---------RAEDSPASTPRE 444
> hsa:88455 ANKRD13A, ANKRD13, NY-REN-25; ankyrin repeat domain
13A
Length=590
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 35 IENPLFHLENTRMLFGNAKSTTSA 58
IE PLFH+ N R+ FGN ++A
Sbjct 411 IEIPLFHVLNARITFGNVNGCSTA 434
> dre:100093707 wu:fj43a03; zgc:165628
Length=208
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 103 REMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKL 141
R+ G+ +QRE+I +H D + A +H P P L
Sbjct 4 RKQGNPQHLSQREIITPEAEHVDAGLAVADLHSHPHPLL 42
> mmu:68420 Ankrd13a, 1100001D10Rik, ANKRD13, AU046136, MGC118502;
ankyrin repeat domain 13a
Length=588
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 1/42 (2%)
Query 18 ARRVFVLKRSMAAGY-ASIENPLFHLENTRMLFGNAKSTTSA 58
AR +K G+ IE PLFH+ N R+ FGN ++A
Sbjct 393 ARLRDFIKLDFPPGFPVKIEIPLFHVLNARITFGNVNGCSTA 434
> cel:Y47G6A.23 lpd-3; LiPid Depleted family member (lpd-3)
Length=1599
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 30/54 (55%), Gaps = 6/54 (11%)
Query 9 GMPVIEAWKARRVFVLKRSMAAGYASIENPLF-----HLENTRMLFGNAKSTTS 57
GM V +WK +V V+ +S A+I N L L+N+R+L GN ++T+
Sbjct 1103 GMNVQASWKDLQV-VITKSTVDDVAAIVNRLISFIDEQLKNSRILLGNLSASTN 1155
> mmu:11899 Astn1, Astn, GC14, mKIAA0289; astrotactin 1
Length=1302
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 5 SKISGMPVIEAWKARRV-FVLKRSMAAGYASIENPLFHLE 43
S++SG PV++ WK R V + +K + AA + N L L+
Sbjct 806 SEVSGYPVLQHWKVRSVMYHIKLNQAAISQAFSNALHSLD 845
> ath:AT1G03160 FZL; FZL (FZO-LIKE); GTP binding / GTPase/ thiamin-phosphate
diphosphorylase
Length=740
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 5 SKISGMPVIEAWKARRVFVLKRS-MAAGYASIENPL-FHLENTRMLFGNAKSTTSAVFAR 62
S+++ + + WK + VF+L +S + +E + F ENTR L V AR
Sbjct 464 SEVAFLRYTQQWKKKFVFILNKSDIYRDARELEEAISFVKENTRKLLNTENVILYPVSAR 523
Query 63 VNARAEQMPPS-AARDDLE 80
A+ S RDDLE
Sbjct 524 SALEAKLSTASLVGRDDLE 542
> hsa:460 ASTN1, ASTN, KIAA1747; astrotactin 1
Length=1294
Score = 28.5 bits (62), Expect = 8.3, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 5 SKISGMPVIEAWKARRV-FVLKRSMAAGYASIENPLFHLE 43
S++SG PV++ WK R V + +K + A ++ N L L+
Sbjct 798 SEVSGYPVLQHWKVRSVMYHIKLNQVAISQALSNALHSLD 837
> tgo:TGME49_058160 hypothetical protein
Length=508
Score = 28.1 bits (61), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 5/46 (10%)
Query 59 VFARVNARAEQMPPSAARDDLESCG----GKFIGLRMGEEAEVLGG 100
VF+R+ AR+EQ+ P AR L +CG + R G + +VLGG
Sbjct 354 VFSRLPARSEQVCPD-ARAALATCGLTQNSRGSPKRSGGQGDVLGG 398
Lambda K H
0.319 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40