bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2668_orf2
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
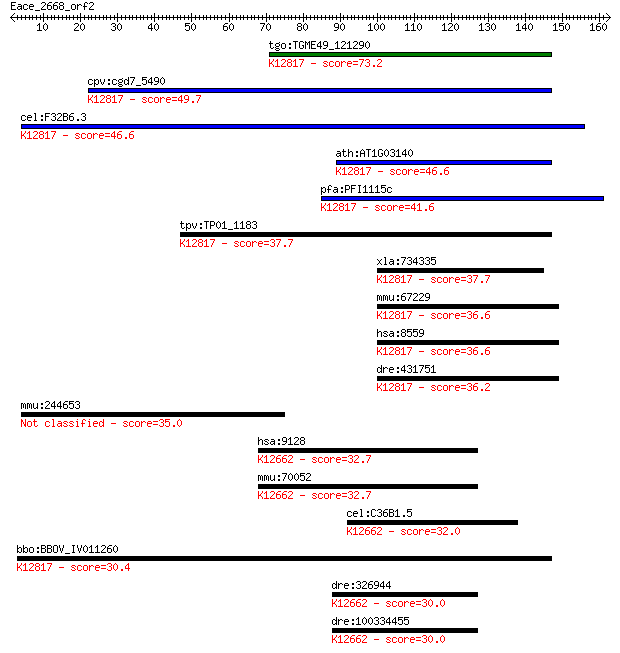
tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K128... 73.2 3e-13
cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-spli... 49.7 5e-06
cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing fac... 46.6 4e-05
ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre... 46.6 4e-05
pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mR... 41.6 0.001
tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRN... 37.7 0.017
xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor ... 37.7 0.018
mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing fac... 36.6 0.037
hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA proce... 36.6 0.037
dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor... 36.2 0.048
mmu:244653 Hydin, 1700034M11Rik, 4930545D19Rik, 4932703P14, A8... 35.0 0.10
hsa:9128 PRPF4, HPRP4, HPRP4P, PRP4, Prp4p; PRP4 pre-mRNA proc... 32.7 0.50
mmu:70052 Prpf4, 1600015H11Rik, AI874830, AW047464, MGC117717,... 32.7 0.51
cel:C36B1.5 prp-4; yeast PRP (splicing factor) related family ... 32.0 0.85
bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K128... 30.4 2.4
dre:326944 prpf4, mg:ab03a02, zgc:65943; PRP4 pre-mRNA process... 30.0 3.1
dre:100334455 PRP4 pre-mRNA processing factor 4 homolog; K1266... 30.0 3.1
> tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K12817
pre-mRNA-splicing factor 18
Length=373
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/76 (46%), Positives = 51/76 (67%), Gaps = 2/76 (2%)
Query 71 SSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKLE 130
S+E + AD D +EE PP+ EIF++LR++K+P++LFGE+ W+RY RLC+LE
Sbjct 24 SAEHAESAADVHDLLN--DEETEPPLPKQEIFKKLRKMKEPVTLFGESDWQRYTRLCELE 81
Query 131 LQAIDDETTEGQKNVF 146
L +DE GQ+N F
Sbjct 82 LLHHEDELMGGQRNAF 97
> cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-splicing
factor 18
Length=337
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 68/134 (50%), Gaps = 9/134 (6%)
Query 22 YILQKDLQQKLREQKDREQEETANKKRKTEL---QQLAELQERLKARAFQPHSSEAEATV 78
++ Q ++Q+ R++ R Q E KK K E ++R+K + SS + +
Sbjct 13 WVSQSEVQEHERQEYIRRQRERDLKKVKKENIKNSHCITSEDRIKKSNHENSSSVIDLSN 72
Query 79 ADA---GDTAEGREEELVP--PVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKLELQA 133
+D + E E L P+ E +RLR L +PI+LFGE +RY+RL +LE +
Sbjct 73 SDCKIISNKLENGSECLTSSLPIQNEETIKRLRLLGEPITLFGEDDNERYNRLRRLEFKG 132
Query 134 -IDDETTEGQKNVF 146
++E GQ+N+F
Sbjct 133 RTNEEMNIGQQNIF 146
> cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing factor
18
Length=352
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 50/154 (32%), Positives = 77/154 (50%), Gaps = 23/154 (14%)
Query 4 IAKQKQQINEL-PKAGPGKYILQKDLQQKLREQKDREQEETANKKRKTELQQLAELQERL 62
+AK+++ ++ + K G K++ DL+ K ++ +R+Q+E A+KKRK + + L E R
Sbjct 11 MAKKRKAVSGMEVKEGNAKFVKGADLEMKRNQEYERKQQEIASKKRKVDDEILQESSSRT 70
Query 63 KARAFQPHSSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKR 122
K P E E+ + +E P + EI RLR+ PI LFGET
Sbjct 71 K-----PAPVENESEI-----------DEKTP---MSEIQTRLRQRNHPIMLFGETDIDV 111
Query 123 YDRLCKLELQAIDDETTEGQKNVFH-AMQREGEE 155
RL +LEL D EG +N AM+ G+E
Sbjct 112 RKRLHQLELAQPD--LNEGWENELQTAMKVIGKE 143
> ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre-mRNA-splicing
factor 18
Length=420
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 36/61 (59%), Gaps = 5/61 (8%)
Query 89 EEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL---CKLELQAIDDETTEGQKNV 145
+E L+ P E+ RRLR LKQP++LFGE R DRL K L +D + TEGQ N
Sbjct 99 DENLILPRQ--EVIRRLRFLKQPMTLFGEDDQSRLDRLKYVLKEGLFEVDSDMTEGQTND 156
Query 146 F 146
F
Sbjct 157 F 157
> pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mRNA-splicing
factor 18
Length=343
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 42/76 (55%), Gaps = 2/76 (2%)
Query 85 AEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKN 144
E E + + +I LR+LK+PI LFGET +RY+RL +L++ +E ++N
Sbjct 75 VENEETNIEITLSNKQIIMLLRQLKEPIRLFGETDLQRYNRLKELKMNK--NELKINEQN 132
Query 145 VFHAMQREGEEEEEFD 160
+F + R +E+ D
Sbjct 133 IFGDVLRGRLKEDSLD 148
> tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRNA-splicing
factor 18
Length=327
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 48/100 (48%), Gaps = 5/100 (5%)
Query 47 KRKTELQQLAELQERLKARAFQPHSSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLR 106
KRK E Q+ E Q +LK + + + E +T + V + E+ +RLR
Sbjct 33 KRKLEAQEQLEKQNQLKKKLINDNFKKIEEFYE--TNTNLDSTHDPTQDVSVEEVVKRLR 90
Query 107 RLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKNVF 146
+L+QPI FGE+ +R RL Q D + E +N++
Sbjct 91 KLRQPIVFFGESHKERCRRLFS---QETDIDDLEANQNIY 127
> xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor
18 homolog; K12817 pre-mRNA-splicing factor 18
Length=342
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query 100 EIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKN 144
E+ RRLR +PI LFGET ++ + RL K+E+ A E +G +N
Sbjct 83 EVIRRLRERGEPIRLFGETDYETFQRLRKIEILA--PEVNKGLRN 125
> mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 36.6 bits (83), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Query 100 EIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKNVFHA 148
E+ RRLR +PI LFGET + + RL K+E+ + E +G +N A
Sbjct 83 EVIRRLRERGEPIRLFGETDYDAFQRLRKIEI--LTPEVNKGLRNDLKA 129
> hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA processing
factor 18 homolog (S. cerevisiae); K12817 pre-mRNA-splicing
factor 18
Length=342
Score = 36.6 bits (83), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Query 100 EIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKNVFHA 148
E+ RRLR +PI LFGET + + RL K+E+ + E +G +N A
Sbjct 83 EVIRRLRERGEPIRLFGETDYDAFQRLRKIEI--LTPEVNKGLRNDLKA 129
> dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 36.2 bits (82), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Query 100 EIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKNVFHA 148
E+ RRLR +PI LFGE+ + + RL K+E+ A E +G +N A
Sbjct 83 EVIRRLRERGEPIRLFGESDYDAFQRLRKIEILA--PEVNKGLRNDLKA 129
> mmu:244653 Hydin, 1700034M11Rik, 4930545D19Rik, 4932703P14,
A830061H17, AC069308.21gm4, hy-3, hy3, hyrh; hydrocephalus inducing
Length=5154
Score = 35.0 bits (79), Expect = 0.10, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 42/80 (52%), Gaps = 9/80 (11%)
Query 4 IAKQKQQINELPK------AGPGKYILQKDLQQKLREQKDREQEETANKKRKTELQQLAE 57
+AK+K+++ L + K +D++Q LRE+K RE E A + ++ +LQQ E
Sbjct 2330 LAKEKERLQTLDEDEYDALTAEEKVAFDRDVRQALRERKKRELERLAKEMQEKKLQQELE 2389
Query 58 LQ---ERLKARAFQPHSSEA 74
Q + LK + +P + A
Sbjct 2390 RQKEEDELKRKVKRPKAGPA 2409
> hsa:9128 PRPF4, HPRP4, HPRP4P, PRP4, Prp4p; PRP4 pre-mRNA processing
factor 4 homolog (yeast); K12662 U4/U6 small nuclear
ribonucleoprotein PRP4
Length=522
Score = 32.7 bits (73), Expect = 0.50, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 1/59 (1%)
Query 68 QPHSSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL 126
+ H SE +A V A R ++ D E+ LR L +PI+LFGE P +R +RL
Sbjct 76 EEHISERQAEVL-AEFERRKRARQINVSTDDSEVKACLRALGEPITLFGEGPAERRERL 133
> mmu:70052 Prpf4, 1600015H11Rik, AI874830, AW047464, MGC117717,
bN189G18.1; PRP4 pre-mRNA processing factor 4 homolog (yeast);
K12662 U4/U6 small nuclear ribonucleoprotein PRP4
Length=521
Score = 32.7 bits (73), Expect = 0.51, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 1/59 (1%)
Query 68 QPHSSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL 126
+ H SE +A V A R ++ D E+ LR L +PI+LFGE P +R +RL
Sbjct 75 EEHISERQAEVL-AEFERRKRARQINVSTDDSEVKACLRALGEPITLFGEGPAERRERL 132
> cel:C36B1.5 prp-4; yeast PRP (splicing factor) related family
member (prp-4); K12662 U4/U6 small nuclear ribonucleoprotein
PRP4
Length=496
Score = 32.0 bits (71), Expect = 0.85, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 92 LVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDE 137
L P D ++ +LR L QPI LFGE R +RL L +DE
Sbjct 72 LTLPTDDVQVKLKLRALNQPICLFGEDALDRRERLRALLSTMSEDE 117
> bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K12817
pre-mRNA-splicing factor 18
Length=285
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 37/144 (25%), Positives = 71/144 (49%), Gaps = 16/144 (11%)
Query 3 IIAKQKQQINELPKAGPGKYILQKDLQQKLREQKDREQEETANKKRKTELQQLAELQERL 62
+I K+K+++ L K G K+ + D +L E++ + +E+ +R +++++ +
Sbjct 7 VIEKKKKEVQALKKDGQ-KWTRKAD---ELAEERKKAEEDI---ERNLQIKKVKTNDQVK 59
Query 63 KARAFQPHSSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKR 122
K +F + +AEA A T L + EI +LRR K+P ++F ET R
Sbjct 60 KISSFYSVNEDAEAKKHKAYIT------NLFKGIPDSEIINQLRRHKEPATIFAET---R 110
Query 123 YDRLCKLELQAIDDETTEGQKNVF 146
DR+ +L + + Q+N+F
Sbjct 111 EDRIERLYQAQEKENIAKNQQNIF 134
> dre:326944 prpf4, mg:ab03a02, zgc:65943; PRP4 pre-mRNA processing
factor 4 homolog (yeast); K12662 U4/U6 small nuclear ribonucleoprotein
PRP4
Length=507
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 88 REEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL 126
R ++ D E+ LR L +PI+LFGE P +R +RL
Sbjct 80 RARQITVSTDDVEVKACLRALGEPITLFGEGPAERRERL 118
> dre:100334455 PRP4 pre-mRNA processing factor 4 homolog; K12662
U4/U6 small nuclear ribonucleoprotein PRP4
Length=507
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 88 REEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL 126
R ++ D E+ LR L +PI+LFGE P +R +RL
Sbjct 80 RARQITVSTDDVEVKACLRALGEPITLFGEGPAERRERL 118
Lambda K H
0.311 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40