bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2734_orf2
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
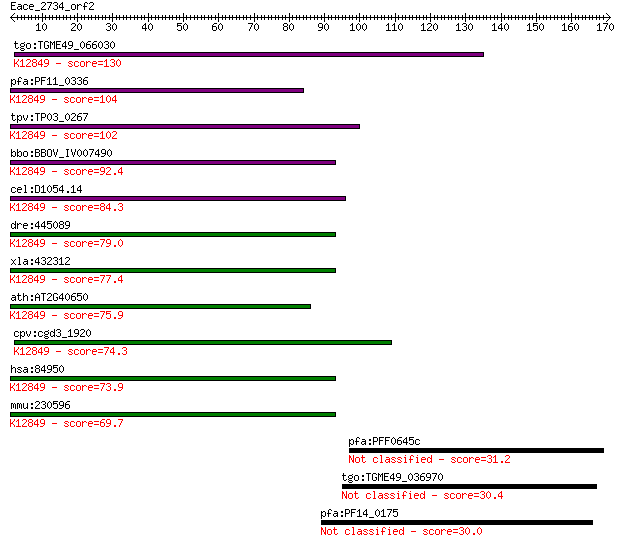
tgo:TGME49_066030 PRP38 family domain-containing protein ; K12... 130 3e-30
pfa:PF11_0336 pre-mRNA splicing factor, putative; K12849 pre-m... 104 1e-22
tpv:TP03_0267 hypothetical protein; K12849 pre-mRNA-splicing f... 102 4e-22
bbo:BBOV_IV007490 23.m06033; pre-mRNA splicing factor; K12849 ... 92.4 7e-19
cel:D1054.14 hypothetical protein; K12849 pre-mRNA-splicing fa... 84.3 2e-16
dre:445089 prpf38a, MGC173831, fa04h11, fc83e08, pmp22b, wu:fa... 79.0 8e-15
xla:432312 prpf38a, MGC116433; PRP38 pre-mRNA processing facto... 77.4 2e-14
ath:AT2G40650 pre-mRNA splicing factor PRP38 family protein; K... 75.9 6e-14
cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family m... 74.3 2e-13
hsa:84950 PRPF38A, FLJ14936, MGC3320, Prp38, RP5-965L7.1; PRP3... 73.9 2e-13
mmu:230596 Prpf38a, 2410002M20Rik; PRP38 pre-mRNA processing f... 69.7 4e-12
pfa:PFF0645c integral membrane protein, putative 31.2 1.6
tgo:TGME49_036970 SNF2 family N-terminal domain containing pro... 30.4 2.9
pfa:PF14_0175 conserved Plasmodium protein, unknown function 30.0 3.7
> tgo:TGME49_066030 PRP38 family domain-containing protein ; K12849
pre-mRNA-splicing factor 38A
Length=524
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 71/138 (51%), Positives = 90/138 (65%), Gaps = 5/138 (3%)
Query 2 FKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRKN 61
FKYLRAV FYLRLV + EVY HLEPLL DYRKLR+R G F++ MD FVD+CLRK
Sbjct 96 FKYLRAVGAFYLRLVGRACEVYTHLEPLLADYRKLRLRLADGKFTIVCMDEFVDDCLRKT 155
Query 62 TLFDIDLPPIVKRKALVQQQLLPPLRLSAL--DVEV---SEQEKSLPLTDSQAHALLLSK 116
D+DLP + KR+ L + LPP RL+AL DVEV S + +++ LL K
Sbjct 156 NFLDVDLPVLPKREVLESEGDLPPARLTALEHDVEVYAPSGVSTAEGVSERDQQLWLLQK 215
Query 117 RLLQQQQHKLQQHVLQQR 134
L+++Q +LQ+ VLQQR
Sbjct 216 EQLRKEQKQLQRRVLQQR 233
> pfa:PF11_0336 pre-mRNA splicing factor, putative; K12849 pre-mRNA-splicing
factor 38A
Length=386
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 47/83 (56%), Positives = 58/83 (69%), Gaps = 0/83 (0%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
DF YLRA+ +FYLRL+ S EVY HLEP+L DYRK+R+R +G F YMDVFVDNCL
Sbjct 95 DFVYLRALGIFYLRLIGKSLEVYNHLEPILFDYRKIRMRLQNGTFEKIYMDVFVDNCLIL 154
Query 61 NTLFDIDLPPIVKRKALVQQQLL 83
N D+D P + KR+ L + LL
Sbjct 155 NNFLDVDFPTLTKRQVLEENNLL 177
> tpv:TP03_0267 hypothetical protein; K12849 pre-mRNA-splicing
factor 38A
Length=193
Score = 102 bits (255), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 49/99 (49%), Positives = 68/99 (68%), Gaps = 1/99 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
D+KYLRA+ ++Y+RLV + EVY LEP+L DYRKLR R+ G++ + YMD FVD+CL
Sbjct 95 DYKYLRALGIYYMRLVGTAVEVYRTLEPILGDYRKLRFRNIDGSYVIKYMDEFVDDCLTN 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALDVEVSEQE 99
T D+DLPP+ KR L + LPP R S L++ ++ E
Sbjct 155 TTYLDVDLPPLSKRMNLEATKALPP-RKSLLNIVKTKDE 192
> bbo:BBOV_IV007490 23.m06033; pre-mRNA splicing factor; K12849
pre-mRNA-splicing factor 38A
Length=202
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 44/92 (47%), Positives = 66/92 (71%), Gaps = 1/92 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
+FKYLRA+ ++Y+RLV + ++Y +LEP+ DYRKLR R+N G++ + +MD FVD+CLR
Sbjct 95 EFKYLRALGIYYMRLVGNAVQIYQNLEPVYADYRKLRFRNNDGSYDIRHMDEFVDDCLRL 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALD 92
++ D+DLP + KR L + + L P R S LD
Sbjct 155 SSYLDVDLPILPKRMILEETKQLGP-RRSLLD 185
> cel:D1054.14 hypothetical protein; K12849 pre-mRNA-splicing
factor 38A
Length=320
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/95 (46%), Positives = 60/95 (63%), Gaps = 1/95 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
+FKY+RA+ YLRL S E+Y +LEPL D+RKLR + G F YMD F+DN LR+
Sbjct 95 EFKYIRALGAMYLRLTFDSTEIYKYLEPLYNDFRKLRYMNKMGRFEAIYMDDFIDNLLRE 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALDVEV 95
+ DI LP + KR AL + +LP + S LD ++
Sbjct 155 DRYCDIQLPRLQKRWALEEVDMLPSYK-SLLDGDL 188
> dre:445089 prpf38a, MGC173831, fa04h11, fc83e08, pmp22b, wu:fa04h11,
wu:fc83e08, zgc:92059; PRP38 pre-mRNA processing factor
38 (yeast) domain containing A; K12849 pre-mRNA-splicing
factor 38A
Length=313
Score = 79.0 bits (193), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 59/92 (64%), Gaps = 1/92 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
DFKY+R + Y+RL S + Y +LEPL DYRK++ ++ +G F L ++D F+D L
Sbjct 95 DFKYVRLLGAMYMRLTGTSVDCYKYLEPLYNDYRKIKSQNRNGEFELMHVDEFIDELLHA 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALD 92
+ DI LP + KR+ L + +LL P R+SAL+
Sbjct 155 ERMCDIILPRLQKRQVLEEAELLDP-RISALE 185
> xla:432312 prpf38a, MGC116433; PRP38 pre-mRNA processing factor
38 domain containing A; K12849 pre-mRNA-splicing factor
38A
Length=312
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 59/92 (64%), Gaps = 1/92 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
DFKY+RA+ Y+RL + + Y +LEPL DYRK++V++ G F L ++D F+D L +
Sbjct 95 DFKYVRALGALYMRLTGTATDCYKYLEPLYNDYRKVKVQNRDGEFELMHVDEFIDQLLHE 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALD 92
+ D+ LP + KR L + + L P R+SAL+
Sbjct 155 ERVCDVILPRLQKRFVLEETEQLDP-RVSALE 185
> ath:AT2G40650 pre-mRNA splicing factor PRP38 family protein;
K12849 pre-mRNA-splicing factor 38A
Length=355
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 38/85 (44%), Positives = 53/85 (62%), Gaps = 0/85 (0%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
D+KY+R + FYLRL +VY +LEPL DYRK+R + + G FSLT++D ++ L K
Sbjct 95 DYKYVRILGAFYLRLTGTDVDVYRYLEPLYNDYRKVRQKLSDGKFSLTHVDEVIEELLTK 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPP 85
+ DI +P + KR L Q LL P
Sbjct 155 DYSCDIAMPRLKKRWTLEQNGLLEP 179
> cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family
member ; K12849 pre-mRNA-splicing factor 38A
Length=261
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 62/107 (57%), Gaps = 0/107 (0%)
Query 2 FKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRKN 61
FKYL A+ + YLRL +S ++ +E L DYR++R+R+ G+F + ++D V+ CL +
Sbjct 89 FKYLTALGIVYLRLTESSIVIHKSIEHLYQDYRRIRIRNLDGSFDIIHLDELVEICLCEK 148
Query 62 TLFDIDLPPIVKRKALVQQQLLPPLRLSALDVEVSEQEKSLPLTDSQ 108
D+D P I KR LV+Q L R S L+ S S P++ ++
Sbjct 149 KFLDLDFPYIHKRAVLVKQGHLKFPRKSMLEYVPSNSGGSSPISKTE 195
> hsa:84950 PRPF38A, FLJ14936, MGC3320, Prp38, RP5-965L7.1; PRP38
pre-mRNA processing factor 38 (yeast) domain containing
A; K12849 pre-mRNA-splicing factor 38A
Length=312
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 57/92 (61%), Gaps = 1/92 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
DFKY+R + Y+RL + + Y +LEPL DYRK++ ++ +G F L ++D F+D L
Sbjct 95 DFKYVRMLGALYMRLTGTAIDCYKYLEPLYNDYRKIKSQNRNGEFELMHVDEFIDELLHS 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALD 92
+ DI LP + KR L + + L P R+SAL+
Sbjct 155 ERVCDIILPRLQKRYVLEEAEQLEP-RVSALE 185
> mmu:230596 Prpf38a, 2410002M20Rik; PRP38 pre-mRNA processing
factor 38 (yeast) domain containing A; K12849 pre-mRNA-splicing
factor 38A
Length=312
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 56/92 (60%), Gaps = 1/92 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
DFKY+R + Y+RL + + Y +LEPL DYRK++ ++ +G F L ++D F+ L
Sbjct 95 DFKYVRMLGALYMRLTGTAIDCYKYLEPLYNDYRKIKSQNRNGEFVLMHVDEFIYELLHS 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALD 92
+ DI LP + KR L + + L P R+SAL+
Sbjct 155 ERVCDIILPRLQKRYVLEEAEQLEP-RVSALE 185
> pfa:PFF0645c integral membrane protein, putative
Length=1422
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 43/82 (52%), Gaps = 10/82 (12%)
Query 97 EQEKSLPLTDSQAHALLLSKRLLQQQQHKLQQHVLQQRNRQLQQQKEIEEERQM------ 150
E++K L Q H +L + +Q+++ KL Q + L+Q K+I+EE+ M
Sbjct 1035 EKKKLLYQKKMQEHNMLEQDQKIQEEKKKLLYQKKMQEHNMLEQDKKIQEEKFMEKKQKL 1094
Query 151 --QLIKEETAPDE--VKRRKID 168
Q ++++ DE ++R KI+
Sbjct 1095 TEQFLEQKQKEDEQILERNKIE 1116
> tgo:TGME49_036970 SNF2 family N-terminal domain containing protein
(EC:3.6.3.8 3.6.3.14)
Length=2556
Score = 30.4 bits (67), Expect = 2.9, Method: Composition-based stats.
Identities = 20/78 (25%), Positives = 37/78 (47%), Gaps = 6/78 (7%)
Query 95 VSEQEKSLPLTDSQAHALLLSKRLLQQQQHKLQQHVLQQRNRQ------LQQQKEIEEER 148
V EQ+ LT + L ++ L+QQQ +L + + +Q + ++ EEER
Sbjct 1734 VEEQQTRRRLTKEEKEQEKLQQKELRQQQKELHHDLDASKRKQENQQRRQECRQRAEEER 1793
Query 149 QMQLIKEETAPDEVKRRK 166
Q Q K++ + K+ +
Sbjct 1794 QFQEAKQQIDSQDAKQEE 1811
> pfa:PF14_0175 conserved Plasmodium protein, unknown function
Length=4662
Score = 30.0 bits (66), Expect = 3.7, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 5/82 (6%)
Query 89 SALDVEVSEQEKSLPLTDSQAHALLLSKRLLQQ---QQHKLQQHVLQQRNRQLQ--QQKE 143
S D+ E+ + L + L + LQQ QQ +LQQ LQQ Q + QQ+
Sbjct 4200 STHDMNQQERLQQERLQQERLQQERLQQERLQQERLQQERLQQERLQQERLQQERLQQER 4259
Query 144 IEEERQMQLIKEETAPDEVKRR 165
+++ER Q +++ E+ ++
Sbjct 4260 LQQERLQQKWEQQKIQQELYQK 4281
Lambda K H
0.321 0.136 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4287611064
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40