bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2735_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
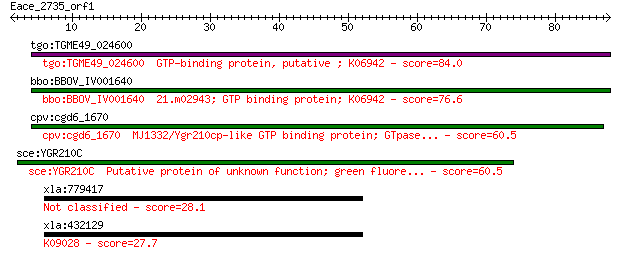
tgo:TGME49_024600 GTP-binding protein, putative ; K06942 84.0 1e-16
bbo:BBOV_IV001640 21.m02943; GTP binding protein; K06942 76.6 2e-14
cpv:cgd6_1670 MJ1332/Ygr210cp-like GTP binding protein; GTpase... 60.5 1e-09
sce:YGR210C Putative protein of unknown function; green fluore... 60.5 1e-09
xla:779417 junb, MGC154397; jun B proto-oncogene 28.1 6.5
xla:432129 junb, MGC81322; jun B proto-oncogene; K09028 transc... 27.7 8.9
> tgo:TGME49_024600 GTP-binding protein, putative ; K06942
Length=451
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 39/84 (46%), Positives = 58/84 (69%), Gaps = 2/84 (2%)
Query 4 RLEKVRDLMLFRHGSTGIAEALRIAVEVLGQRIAGYPVKSLQLTATDKKGRGPFIECLLV 63
RLEK++D++++R+GS+G+ EA+R AV LG R+ YPV+ T + KG + ECLLV
Sbjct 340 RLEKIKDMVIYRYGSSGVHEAIRAAVTRLGNRVPVYPVRHWNTTGS--KGANLYAECLLV 397
Query 64 KKGTTIGDFASLLHPAIGRNFISA 87
+ T + DFA +LH A+ RNF+ A
Sbjct 398 PRNTRMRDFAGMLHVAMERNFLYA 421
> bbo:BBOV_IV001640 21.m02943; GTP binding protein; K06942
Length=423
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 40/84 (47%), Positives = 52/84 (61%), Gaps = 1/84 (1%)
Query 4 RLEKVRDLMLFRHGSTGIAEALRIAVEVLGQRIAGYPVKSLQLTATDKKGRGPFIECLLV 63
RL +RDL+L+R GSTG+ AL A + G I YPVK+L+ D+ F EC+ V
Sbjct 309 RLNNIRDLVLYRFGSTGVTSALNEATKAAGV-ICVYPVKNLKTFGDDESSSKAFRECITV 367
Query 64 KKGTTIGDFASLLHPAIGRNFISA 87
GTT+ +FAS+LH IGRNF A
Sbjct 368 MPGTTVREFASMLHFEIGRNFHKA 391
> cpv:cgd6_1670 MJ1332/Ygr210cp-like GTP binding protein; GTpase
OBG family plus RNA binding domain TGS ; K06942
Length=443
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 31/86 (36%), Positives = 51/86 (59%), Gaps = 4/86 (4%)
Query 4 RLEKVRDLMLFRHGSTGIAEALRIAVEVLGQRIAGYPVKSLQLTAT---DKKGRGPFIEC 60
RLE ++D++LFRHG TG+ +A+ AV++LG I YPVK+++ D+ F +C
Sbjct 326 RLENIKDMVLFRHGVTGVQDAVNKAVDLLG-LIPIYPVKNIKTFTNNLHDETNNSAFQDC 384
Query 61 LLVKKGTTIGDFASLLHPAIGRNFIS 86
+LV GT + LH + +N ++
Sbjct 385 ILVPNGTMVKTLLKSLHIDLEKNLVN 410
> sce:YGR210C Putative protein of unknown function; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm;
K06942
Length=411
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Query 2 FHRLEKVRDLMLFRHGSTGIAEALRIAVEVLGQRIAGYPVKSLQLTATDKKGRGPFIECL 61
+R+E +RDL+L+R GSTG+ + L+ A ++LG I Y VK++Q T T G F +C
Sbjct 298 LNRIENIRDLVLYRFGSTGVVQVLQAATDILG-LIPVYTVKNIQ-TFTGGNGTNVFRDCF 355
Query 62 LVKKGTTIGDFA 73
LVK+GT +G A
Sbjct 356 LVKRGTPVGKVA 367
> xla:779417 junb, MGC154397; jun B proto-oncogene
Length=295
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 6 EKVRDLMLFRHGSTGIAEALRIAVEVLGQRIAGYPVKSLQLTATDK 51
EKVRDL +G +G A ALR VE L R+ + L T K
Sbjct 245 EKVRDLKNENNGLSGTAGALREQVEQLKVRVREHARHGCHLLLTGK 290
> xla:432129 junb, MGC81322; jun B proto-oncogene; K09028 transcription
factor jun-B
Length=302
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 6 EKVRDLMLFRHGSTGIAEALRIAVEVLGQRIAGYPVKSLQLTATDK 51
EKVR+L G +G A ALR VE L R+ + QL T K
Sbjct 252 EKVRELKNENSGLSGTAGALREQVEQLKVRVREHARHGCQLLLTGK 297
Lambda K H
0.325 0.142 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026251472
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40