bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2743_orf1
Length=136
Score E
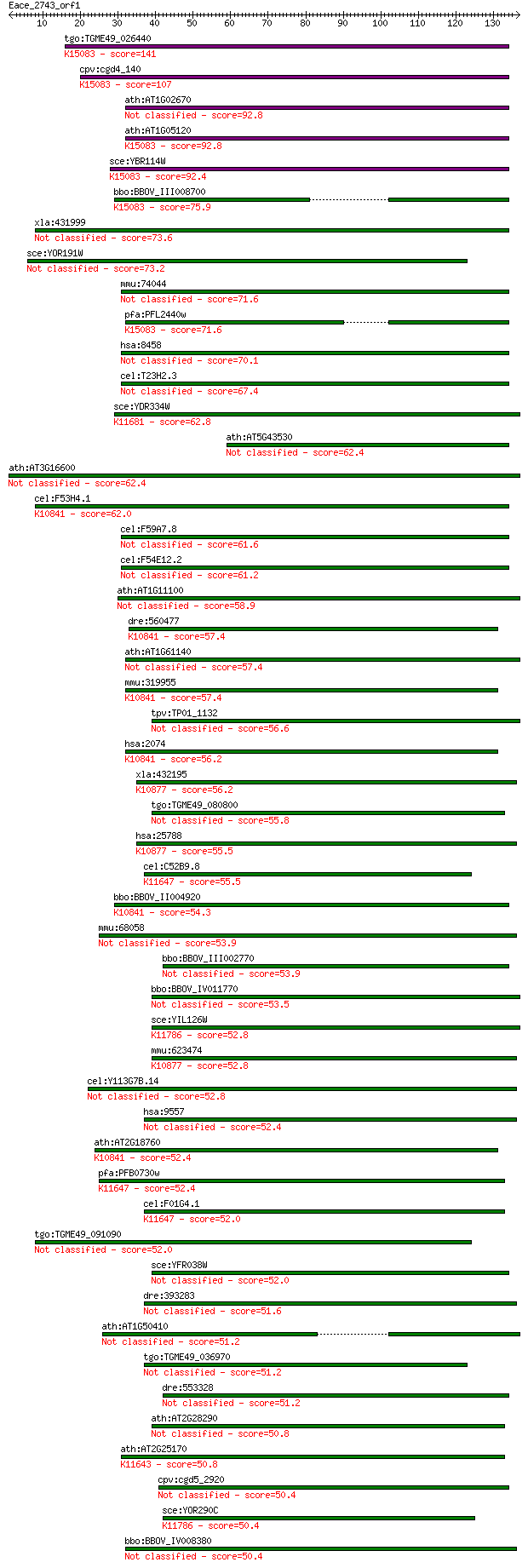
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_026440 SNF2/RAD54 helicase family protein (EC:2.7.1... 141 6e-34
cpv:cgd4_140 Swi2/Snf2 ATpase,Rad16 ortholog ; K15083 DNA repa... 107 8e-24
ath:AT1G02670 DNA repair protein, putative 92.8 3e-19
ath:AT1G05120 SNF2 domain-containing protein / helicase domain... 92.8 3e-19
sce:YBR114W RAD16, PSO5; Protein that recognizes and binds dam... 92.4 3e-19
bbo:BBOV_III008700 17.m10681; DNA repair protein rhp16; K15083... 75.9 4e-14
xla:431999 ttf2, MGC81081; transcription termination factor, R... 73.6 2e-13
sce:YOR191W ULS1, DIS1, RIS1, TID4; RING finger protein involv... 73.2 2e-13
mmu:74044 Ttf2, 4632434F22Rik, 8030447N19, AV218430; transcrip... 71.6 6e-13
pfa:PFL2440w DNA repair protein rhp16, putative; K15083 DNA re... 71.6 6e-13
hsa:8458 TTF2, HuF2; transcription termination factor, RNA pol... 70.1 2e-12
cel:T23H2.3 hypothetical protein 67.4 1e-11
sce:YDR334W SWR1; Swi2/Snf2-related ATPase that is the structu... 62.8 3e-10
ath:AT5G43530 SNF2 domain-containing protein / helicase domain... 62.4 4e-10
ath:AT3G16600 SNF2 domain-containing protein / helicase domain... 62.4 4e-10
cel:F53H4.1 csb-1; human CSB (Cockayne Syndrome B) homolog fam... 62.0 5e-10
cel:F59A7.8 hypothetical protein 61.6 6e-10
cel:F54E12.2 hypothetical protein 61.2 9e-10
ath:AT1G11100 SNF2 domain-containing protein / helicase domain... 58.9 5e-09
dre:560477 ercc6; excision repair cross-complementing rodent r... 57.4 1e-08
ath:AT1G61140 EDA16; EDA16 (embryo sac development arrest 16);... 57.4 1e-08
mmu:319955 Ercc6, 4732403I04, C130058G22Rik, CSB; excision rep... 57.4 1e-08
tpv:TP01_1132 ATP-dependent helicase 56.6 2e-08
hsa:2074 ERCC6, ARMD5, CKN2, COFS, COFS1, CSB, RAD26; excision... 56.2 3e-08
xla:432195 rad54b, MGC81308, fsbp, rdh54; RAD54 homolog B; K10... 56.2 3e-08
tgo:TGME49_080800 SNF2 family N-terminal domain-containing pro... 55.8 4e-08
hsa:25788 RAD54B, FSBP, RDH54; RAD54 homolog B (S. cerevisiae)... 55.5 5e-08
cel:C52B9.8 hypothetical protein; K11647 SWI/SNF-related matri... 55.5 5e-08
bbo:BBOV_II004920 18.m06409; SNF2 domain-containing protein / ... 54.3 1e-07
mmu:68058 Chd1l, 4432404A22Rik, Alc1, Snf2p; chromodomain heli... 53.9 1e-07
bbo:BBOV_III002770 17.m07263; SNF2 family N-terminal domain co... 53.9 1e-07
bbo:BBOV_IV011770 23.m06400; snf2-related chromatin remodeling... 53.5 2e-07
sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin ... 52.8 3e-07
mmu:623474 Rad54b, E130016E03Rik, Fsbp, MGC67261; RAD54 homolo... 52.8 3e-07
cel:Y113G7B.14 hypothetical protein 52.8 3e-07
hsa:9557 CHD1L, ALC1, CHDL, FLJ22530; chromodomain helicase DN... 52.4 4e-07
ath:AT2G18760 CHR8; CHR8 (chromatin remodeling 8); ATP binding... 52.4 4e-07
pfa:PFB0730w DEAD/DEAH box helicase, putative; K11647 SWI/SNF-... 52.4 4e-07
cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4)... 52.0 5e-07
tgo:TGME49_091090 SNF2 family N-terminal domain-containing pro... 52.0 5e-07
sce:YFR038W IRC5; Irc5p (EC:3.6.1.-) 52.0 5e-07
dre:393283 chd1l, MGC56084, zgc:56084; chromodomain helicase D... 51.6 7e-07
ath:AT1G50410 SNF2 domain-containing protein / helicase domain... 51.2 8e-07
tgo:TGME49_036970 SNF2 family N-terminal domain containing pro... 51.2 9e-07
dre:553328 hells, cb65, im:6911667, pasg, sb:cb65, sb:cb749; h... 51.2 1e-06
ath:AT2G28290 SYD; SYD (SPLAYED); ATPase/ chromatin binding 50.8 1e-06
ath:AT2G25170 PKL; PKL (PICKLE); ATPase/ DNA binding / DNA hel... 50.8 1e-06
cpv:cgd5_2920 hypothetical protein 50.4 1e-06
sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit of... 50.4 1e-06
bbo:BBOV_IV008380 23.m05834; SNF2 helicase (EC:3.6.1.-) 50.4 2e-06
> tgo:TGME49_026440 SNF2/RAD54 helicase family protein (EC:2.7.11.1
6.1.1.11); K15083 DNA repair protein RAD16
Length=1667
Score = 141 bits (355), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 79/125 (63%), Positives = 95/125 (76%), Gaps = 7/125 (5%)
Query 16 MLSGLTKREIKGRVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQI 75
++ L KR+I+GR +P LLLPLLPFQEEGLWWL +QE S ++GGILADEMGMGKTIQI
Sbjct 241 VMRNLVKRQIEGRRPAPPDLLLPLLPFQEEGLWWLCRQEQSEVRGGILADEMGMGKTIQI 300
Query 76 ISLLLSRPLPTIPESAPPLVRA-------CVCSTLIVTPLAALLQWKSELDKFVAPGRLS 128
ISL+L+RP P +P + P + V TL+VTPLAALLQWK EL+KFV PGRLS
Sbjct 301 ISLILARPFPPLPRALRPEDSSRERSSLPRVGQTLVVTPLAALLQWKGELEKFVRPGRLS 360
Query 129 VLIYH 133
VL+YH
Sbjct 361 VLVYH 365
> cpv:cgd4_140 Swi2/Snf2 ATpase,Rad16 ortholog ; K15083 DNA repair
protein RAD16
Length=1278
Score = 107 bits (268), Expect = 8e-24, Method: Composition-based stats.
Identities = 55/114 (48%), Positives = 69/114 (60%), Gaps = 2/114 (1%)
Query 20 LTKREIKGRVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLL 79
L R I +P L LL FQ+EGL WL QE S +GGILADEMGMGKTIQ ISL+
Sbjct 166 LISRSIVEVKPTPIKLTYELLQFQKEGLAWLCNQEKSTARGGILADEMGMGKTIQTISLI 225
Query 80 LSRPLPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
L +P + A + L++ P+AA+LQWK E+++F PG L V IYH
Sbjct 226 LEHDIPPVTNKAEK--GEVIGKNLVIAPVAAVLQWKQEIERFTKPGSLKVHIYH 277
> ath:AT1G02670 DNA repair protein, putative
Length=678
Score = 92.8 bits (229), Expect = 3e-19, Method: Composition-based stats.
Identities = 50/102 (49%), Positives = 64/102 (62%), Gaps = 5/102 (4%)
Query 32 PEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESA 91
P L++PLL +Q+E L W T QE S ++GGILADEMGMGKTIQ ISL+L+R +S
Sbjct 127 PLDLIIPLLKYQKEFLAWATIQELSAVRGGILADEMGMGKTIQAISLVLARREVDRAKS- 185
Query 92 PPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
R V TL++ P AL QW E+ + +PG VL YH
Sbjct 186 ----REAVGHTLVLVPPVALSQWLDEISRLTSPGSTRVLQYH 223
> ath:AT1G05120 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein;
K15083 DNA repair protein RAD16
Length=833
Score = 92.8 bits (229), Expect = 3e-19, Method: Composition-based stats.
Identities = 49/102 (48%), Positives = 67/102 (65%), Gaps = 6/102 (5%)
Query 32 PEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESA 91
P L++PLL +Q+E L W T+QE S + GGILADEMGMGKTIQ ISL+L+R + +
Sbjct 132 PSDLIMPLLRYQKEFLAWATKQEQS-VAGGILADEMGMGKTIQAISLVLAR--REVDRAQ 188
Query 92 PPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
C TL++ PL A+ QW +E+ +F +PG VL+YH
Sbjct 189 FGEAAGC---TLVLCPLVAVSQWLNEIARFTSPGSTKVLVYH 227
> sce:YBR114W RAD16, PSO5; Protein that recognizes and binds damaged
DNA in an ATP-dependent manner (with Rad7p) during nucleotide
excision repair; subunit of Nucleotide Excision Repair
Factor 4 (NEF4) and the Elongin-Cullin-Socs (ECS) ligase
complex (EC:3.6.1.-); K15083 DNA repair protein RAD16
Length=790
Score = 92.4 bits (228), Expect = 3e-19, Method: Composition-based stats.
Identities = 49/106 (46%), Positives = 65/106 (61%), Gaps = 15/106 (14%)
Query 28 RVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTI 87
R P+G+ + LLPFQ EGL WL QE S GG+LADEMGMGKTIQ I+LL++ L
Sbjct 173 RSKQPDGMTIKLLPFQLEGLHWLISQEESIYAGGVLADEMGMGKTIQTIALLMN-DLTKS 231
Query 88 PESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
P +L+V P AL+QWK+E+++ G+L + IYH
Sbjct 232 P-------------SLVVAPTVALMQWKNEIEQHTK-GQLKIYIYH 263
> bbo:BBOV_III008700 17.m10681; DNA repair protein rhp16; K15083
DNA repair protein RAD16
Length=1289
Score = 75.9 bits (185), Expect = 4e-14, Method: Composition-based stats.
Identities = 33/52 (63%), Positives = 41/52 (78%), Gaps = 0/52 (0%)
Query 29 VASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLL 80
V P LL+PLLPFQ++G+ W+ QQE ++GGILADEMGMGKTIQ I LL+
Sbjct 131 VHQPSQLLIPLLPFQKDGVAWMQQQEMGPVRGGILADEMGMGKTIQTIGLLV 182
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/32 (59%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 102 TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
TLI++PLAALLQW +E+ V G +SVL+YH
Sbjct 274 TLIISPLAALLQWYNEIKTKVEDGFISVLLYH 305
> xla:431999 ttf2, MGC81081; transcription termination factor,
RNA polymerase II
Length=1187
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 49/136 (36%), Positives = 70/136 (51%), Gaps = 10/136 (7%)
Query 8 ATTEAAEGMLSGLTKR-EIKGRVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADE 66
AT+EA E + L + V P GL +PLL Q++ L WL +E +GGILAD+
Sbjct 561 ATSEAIEHLHKSLESCPSPENTVEDPAGLKVPLLLHQKQALAWLRWRENQTPRGGILADD 620
Query 67 MGMGKTIQIISLLLSRPLPTIPESAPPLVR---------ACVCSTLIVTPLAALLQWKSE 117
MG+GKT+ +++L+L + E L TLIV P + + WK E
Sbjct 621 MGLGKTLTMVALILMQKQRQNREQEKKLEEWISKTDSTLVVTRGTLIVCPASLVHHWKKE 680
Query 118 LDKFVAPGRLSVLIYH 133
++K VA RL V +YH
Sbjct 681 VEKRVAGSRLKVYLYH 696
> sce:YOR191W ULS1, DIS1, RIS1, TID4; RING finger protein involved
in proteolytic control of sumoylated substrates; interacts
with SUMO (Smt3p); member of the SWI/SNF family of DNA-dependent
ATPases; plays a role in antagonizing silencing during
mating-type switching (EC:3.6.1.-)
Length=1619
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 49/120 (40%), Positives = 64/120 (53%), Gaps = 14/120 (11%)
Query 6 AAATTEAAEGMLSGLTKRE--IKGRVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGIL 63
AA E +L + + E I G +PE + + LL Q GL WL Q E S KGG+L
Sbjct 908 AAEDQEQIRALLENVKQSESIIDGEALTPEDMTVNLLKHQRLGLHWLLQVENSAKKGGLL 967
Query 64 ADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCST-LIVTPLAALLQWKSELDKFV 122
AD+MG+GKTIQ I+L+L+ ES C T LIV P++ L WK EL+ V
Sbjct 968 ADDMGLGKTIQAIALMLAN---RSEESK--------CKTNLIVAPVSVLRVWKGELETKV 1016
> mmu:74044 Ttf2, 4632434F22Rik, 8030447N19, AV218430; transcription
termination factor, RNA polymerase II
Length=1138
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 41/116 (35%), Positives = 66/116 (56%), Gaps = 15/116 (12%)
Query 31 SPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLL---------- 80
P GL +PLL Q++ L WL +E+ +GGILAD+MG+GKT+ +I+L+L
Sbjct 538 DPAGLKVPLLLHQKQALAWLLWRESQKPQGGILADDMGLGKTLTMIALILTKKNQQKSKE 597
Query 81 ---SRPLPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
S P+ + ++ + + TLIV P + + WK+E++K V RL + +YH
Sbjct 598 KERSEPVTWLSKNDSSVFTS--SGTLIVCPASLIHHWKNEVEKRVTSNRLRIYLYH 651
> pfa:PFL2440w DNA repair protein rhp16, putative; K15083 DNA
repair protein RAD16
Length=1647
Score = 71.6 bits (174), Expect = 6e-13, Method: Composition-based stats.
Identities = 32/58 (55%), Positives = 44/58 (75%), Gaps = 0/58 (0%)
Query 32 PEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPE 89
P+ L LL +Q+EG++W+ QE S++KGGILADEMGMGKTIQ I+L+L + L + E
Sbjct 219 PKELKYDLLQYQKEGIYWMINQEMSNVKGGILADEMGMGKTIQAITLILCQKLNKLKE 276
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 18/32 (56%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 102 TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
TLI+ P+AA++QWKSE++KFV L+V +YH
Sbjct 443 TLIIAPVAAVMQWKSEIEKFVDENILNVYVYH 474
> hsa:8458 TTF2, HuF2; transcription termination factor, RNA polymerase
II
Length=1162
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 64/114 (56%), Gaps = 11/114 (9%)
Query 31 SPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPES 90
P GL +PLL Q++ L WL +E+ +GGILAD+MG+GKT+ +I+L+L++ E
Sbjct 562 DPAGLKVPLLLHQKQALAWLLWRESQKPQGGILADDMGLGKTLTMIALILTQKNQEKKEE 621
Query 91 APP------LVRACVCS-----TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
L + C TLI+ P + + WK+E++K V +L V +YH
Sbjct 622 KEKSTALTWLSKDDSCDFTSHGTLIICPASLIHHWKNEVEKRVNSNKLRVYLYH 675
> cel:T23H2.3 hypothetical protein
Length=1001
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 39/117 (33%), Positives = 66/117 (56%), Gaps = 14/117 (11%)
Query 31 SPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLL-----SRPLP 85
+P+GLL+ L+P Q+ GL WL +E GGILAD+MG+GKT+ ++SL++ R
Sbjct 364 TPDGLLVELMPHQKAGLRWLVWREGQPHSGGILADDMGLGKTLSMLSLIVHQKAARRARK 423
Query 86 TIPESAPPLVRACVCS---------TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
++A + V TLI+ P + + QW++E+++ + LSV ++H
Sbjct 424 ESGDNAADKEKRRVAKEEGLYPSNGTLIIAPASLIHQWEAEINRRLESDLLSVFMFH 480
> sce:YDR334W SWR1; Swi2/Snf2-related ATPase that is the structural
component of the SWR1 complex, which exchanges histone
variant H2AZ (Htz1p) for chromatin-bound histone H2A (EC:3.6.1.-);
K11681 helicase SWR1 [EC:3.6.4.12]
Length=1514
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 46/108 (42%), Positives = 56/108 (51%), Gaps = 13/108 (12%)
Query 29 VASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIP 88
V P L L +Q++GL WL +H G ILADEMG+GKTIQ ISLL L
Sbjct 686 VPVPSLLRGNLRTYQKQGLNWLASLYNNHTNG-ILADEMGLGKTIQTISLLAY--LACEK 742
Query 89 ESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRDP 136
E+ P LIV P + LL W+ E +F APG VL Y+ P
Sbjct 743 ENWGP--------HLIVVPTSVLLNWEMEFKRF-APG-FKVLTYYGSP 780
> ath:AT5G43530 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=1277
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 49/94 (52%), Gaps = 19/94 (20%)
Query 59 KGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCS----------------- 101
+GGILAD MG+GKT+ I+L+L+RP PE+ LV
Sbjct 681 RGGILADAMGLGKTVMTIALILARPGRGNPENEDVLVADVNADKRNRKEIHMALTTVKAK 740
Query 102 --TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
TLI+ P+A L QWK EL+ P +SVL+Y+
Sbjct 741 GGTLIICPMALLSQWKDELETHSKPDTVSVLVYY 774
> ath:AT3G16600 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=638
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 48/145 (33%), Positives = 76/145 (52%), Gaps = 10/145 (6%)
Query 1 EVQKEAAATTEAAEGMLSGLTKREIKGRVASPEGLL-LPLLPFQEEGLWWLTQQE--ASH 57
EVQ+E E + ++ K P G+L +PL+ Q+ L W+ ++E + H
Sbjct 14 EVQEEKTTVNERVIYQAALQDLKQPKTEKDLPPGVLTVPLMRHQKIALNWMRKKEKRSRH 73
Query 58 IKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCSTLIVTPLAALLQWKSE 117
GGILAD+ G+GKTI ISL+L + L + + + TLIV P + + QW E
Sbjct 74 CLGGILADDQGLGKTISTISLILLQKLKSQSKQRKRKGQNS-GGTLIVCPASVVKQWARE 132
Query 118 LDKFVA-PGRLSVLIYH-----RDP 136
+ + V+ +LSVL++H +DP
Sbjct 133 VKEKVSDEHKLSVLVHHGSHRTKDP 157
> cel:F53H4.1 csb-1; human CSB (Cockayne Syndrome B) homolog family
member (csb-1); K10841 DNA excision repair protein ERCC-6
Length=957
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 46/131 (35%), Positives = 69/131 (52%), Gaps = 14/131 (10%)
Query 8 ATTEAAEGMLSGLTKREIKGRVASP----EGLLLPLLPFQEEGLWWLTQQEASHIKGGIL 63
AT+E + + + E G+ + P G+ L FQ+EG+ WL Q++ H GGIL
Sbjct 162 ATSELVD---TRMVTSEEYGKSSKPWKVNAGVWNKLHKFQQEGVEWL-QKKTDHRSGGIL 217
Query 64 ADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCST-LIVTPLAALLQWKSELDKFV 122
ADEMG+GKTIQ + L S I E+A + T LIV ++ + QW EL+++
Sbjct 218 ADEMGLGKTIQSVVFLRS-----IQETARTHYKTTGLDTALIVCHVSIIAQWIKELNQWF 272
Query 123 APGRLSVLIYH 133
R+ +L H
Sbjct 273 PKARVFLLHSH 283
> cel:F59A7.8 hypothetical protein
Length=518
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 57/103 (55%), Gaps = 8/103 (7%)
Query 31 SPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPES 90
+P+G L+ L+P Q+ GL WL +E+ GGIL +MG+GKT+ +ISL+ + +
Sbjct 136 TPDGFLVDLMPHQKAGLCWLLWRESQPHSGGILGGDMGLGKTLSMISLI-------VHQK 188
Query 91 APPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
A R I P + + W++E+ + + LSVL+YH
Sbjct 189 AARKTRKDAGDDAIA-PESLVHHWEAEIARRLKQDLLSVLVYH 230
> cel:F54E12.2 hypothetical protein
Length=1091
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 38/117 (32%), Positives = 59/117 (50%), Gaps = 14/117 (11%)
Query 31 SPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIP-- 88
+P+G L L+P Q+ GL W+ +E GGILAD+MG+GKT+ +ISL+ +
Sbjct 463 TPKGFKLELMPHQKAGLTWMRWRETQPQPGGILADDMGLGKTLSMISLIAHQKAARRARR 522
Query 89 ----ESAPPLVRACV--------CSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
+ R V TLIV P + + QW +E+D+ + LS ++H
Sbjct 523 EDGNDDKDKEKRKVVKEQGLIPSNGTLIVAPASLIHQWDAEIDRRLDDSVLSTYMFH 579
> ath:AT1G11100 SNF2 domain-containing protein / helicase domain-containing
protein / zinc finger protein-related
Length=1226
Score = 58.9 bits (141), Expect = 5e-09, Method: Composition-based stats.
Identities = 48/144 (33%), Positives = 69/144 (47%), Gaps = 37/144 (25%)
Query 30 ASPEGLL-LPLLPFQEEGLWWLTQQEASHIK--GGILADEMGMGKTIQIISLLLS-RPLP 85
+ P+G+L + LL Q L W++Q+E S GGILAD+ G+GKT+ I+L+L+ R P
Sbjct 532 SPPDGVLAVSLLRHQRIALSWMSQKETSGNPCFGGILADDQGLGKTVSTIALILTERSTP 591
Query 86 TIP-------------------ESAPPLVRACVCS--------TLIVTPLAALLQWKSEL 118
+P + +V +C TLIV P + + QW EL
Sbjct 592 YLPCEEDSKNGGCNQSDHSQVVFNENKVVEDSLCKMRGRPAAGTLIVCPTSLMRQWADEL 651
Query 119 DKFVA-PGRLSVLIYH-----RDP 136
K V LSVL+YH +DP
Sbjct 652 RKKVTLEAHLSVLVYHGCSRTKDP 675
> dre:560477 ercc6; excision repair cross-complementing rodent
repair deficiency, complementation group 6; K10841 DNA excision
repair protein ERCC-6
Length=1390
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 50/104 (48%), Gaps = 7/104 (6%)
Query 33 EGLLLP------LLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPT 86
EG +P L +Q+ G+ W+ + GGIL DEMG+GKTIQII+ L
Sbjct 472 EGFKIPGFLWKKLFKYQQTGVRWMWELHCQQA-GGILGDEMGLGKTIQIIAFLAGLSYSK 530
Query 87 IPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVL 130
+ A + T+IV P + QW E + P R++VL
Sbjct 531 LKTRGSNYRYAGLGPTVIVCPATVMHQWVKEFHTWWPPFRVAVL 574
> ath:AT1G61140 EDA16; EDA16 (embryo sac development arrest 16);
ATP binding / DNA binding / helicase/ nucleic acid binding
/ protein binding / zinc ion binding
Length=1022
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 51/160 (31%), Positives = 69/160 (43%), Gaps = 55/160 (34%)
Query 32 PEGLL-LPLLPFQEEGLWWLTQQEASHI--KGGILADEMGMGKTIQIISLLL---SRPLP 85
P+G+L +PLL Q L W+ Q+E S GGILAD+ G+GKT+ I+L+L S+P
Sbjct 555 PDGVLTVPLLRHQRIALSWMAQKETSGFPCSGGILADDQGLGKTVSTIALILKERSKPAQ 614
Query 86 TIPES---------------AP--PLVRA--------------------------CVCST 102
ES AP P R+ T
Sbjct 615 ACEESTKKEIFDLESETGECAPLKPSGRSKHFEHSQLLSNENKVGGDSVGKVTGRPAAGT 674
Query 103 LIVTPLAALLQWKSELDKFV-APGRLSVLIYH-----RDP 136
L+V P + + QW EL K V + LSVL+YH +DP
Sbjct 675 LVVCPTSVMRQWADELHKKVTSEANLSVLVYHGSSRTKDP 714
> mmu:319955 Ercc6, 4732403I04, C130058G22Rik, CSB; excision repair
cross-complementing rodent repair deficiency, complementation
group 6; K10841 DNA excision repair protein ERCC-6
Length=1481
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 47/99 (47%), Gaps = 1/99 (1%)
Query 32 PEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESA 91
P L L +Q+ G+ WL + GGIL DEMG+GKTIQII+ L I
Sbjct 496 PGFLFKKLFKYQQTGVRWLWELHCQQA-GGILGDEMGLGKTIQIIAFLAGLSYSKIRTRG 554
Query 92 PPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVL 130
+ T+IV P + QW E + P R++VL
Sbjct 555 SNYRFEGLGPTIIVCPTTVMHQWVKEFHTWWPPFRVAVL 593
> tpv:TP01_1132 ATP-dependent helicase
Length=1632
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 40/98 (40%), Positives = 51/98 (52%), Gaps = 13/98 (13%)
Query 39 LLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRAC 98
L P+Q+EGL WL +I G ILADEMG+GKT+Q I LL L + P
Sbjct 698 LRPYQKEGLRWLVSLYERNING-ILADEMGLGKTLQTICLLAY--LACNKGNWGP----- 749
Query 99 VCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRDP 136
+IV P + LL W E +KF PG +L Y+ P
Sbjct 750 ---HIIVVPTSILLNWVMEFNKF-CPG-FKILAYYGTP 782
> hsa:2074 ERCC6, ARMD5, CKN2, COFS, COFS1, CSB, RAD26; excision
repair cross-complementing rodent repair deficiency, complementation
group 6; K10841 DNA excision repair protein ERCC-6
Length=1493
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 34/99 (34%), Positives = 47/99 (47%), Gaps = 1/99 (1%)
Query 32 PEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESA 91
P L L +Q+ G+ WL + GGIL DEMG+GKTIQII+ L I
Sbjct 500 PGFLFKKLFKYQQTGVRWLWELHCQQ-AGGILGDEMGLGKTIQIIAFLAGLSYSKIRTRG 558
Query 92 PPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVL 130
+ T+IV P + QW E + P R+++L
Sbjct 559 SNYRFEGLGPTVIVCPTTVMHQWVKEFHTWWPPFRVAIL 597
> xla:432195 rad54b, MGC81308, fsbp, rdh54; RAD54 homolog B; K10877
DNA repair and recombination protein RAD54B [EC:3.6.4.-]
Length=895
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 57/105 (54%), Gaps = 9/105 (8%)
Query 35 LLLPLLPFQEEGLWWLTQ----QEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPES 90
L + L P Q+EG+ +L + + G ILADEMG+GKT+Q ISL+ + L P
Sbjct 275 LAVHLRPHQKEGILFLYECVMGMRVNERFGAILADEMGLGKTLQCISLIWTL-LRQGPYG 333
Query 91 APPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
A P+++ LIVTP + + W+ E K++ R+ V +D
Sbjct 334 AKPVIK----RALIVTPGSLVKNWRKEFQKWLGTERIRVFAVDQD 374
> tgo:TGME49_080800 SNF2 family N-terminal domain-containing protein
(EC:2.7.11.1 2.7.1.127)
Length=2894
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 52/96 (54%), Gaps = 17/96 (17%)
Query 39 LLPFQEEGLWWLTQQEASHIKG--GILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVR 96
L +Q EG+ WL A H KG GILADEMG+GKT+Q I +LL+R L R
Sbjct 1221 LRTYQSEGVQWLF---ALHDKGLNGILADEMGLGKTLQTI-VLLARLA---------LER 1267
Query 97 ACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIY 132
LIV P + +L W+ E KF PG VL+Y
Sbjct 1268 GVWGPHLIVVPTSVMLNWEREFFKFC-PG-FKVLVY 1301
> hsa:25788 RAD54B, FSBP, RDH54; RAD54 homolog B (S. cerevisiae);
K10877 DNA repair and recombination protein RAD54B [EC:3.6.4.-]
Length=910
Score = 55.5 bits (132), Expect = 5e-08, Method: Composition-based stats.
Identities = 36/105 (34%), Positives = 56/105 (53%), Gaps = 9/105 (8%)
Query 35 LLLPLLPFQEEGLWWLTQ----QEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPES 90
L+ L P Q+EG+ +L + + G ILADEMG+GKT+Q ISL+ + P
Sbjct 292 LVYHLRPHQKEGIIFLYECVMGMRMNGRCGAILADEMGLGKTLQCISLIWTLQCQG-PYG 350
Query 91 APPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
P+++ TLIVTP + + WK E K++ R+ + +D
Sbjct 351 GKPVIK----KTLIVTPGSLVNNWKKEFQKWLGSERIKIFTVDQD 391
> cel:C52B9.8 hypothetical protein; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1336
Score = 55.5 bits (132), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/87 (36%), Positives = 51/87 (58%), Gaps = 11/87 (12%)
Query 37 LPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVR 96
L L P+Q +GL W+ +++ G ILADEMG+GKTIQ I+ + L I +++ P
Sbjct 364 LKLKPYQIKGLEWMVSLFNNNLNG-ILADEMGLGKTIQTIAFITY--LMEIKKTSGPF-- 418
Query 97 ACVCSTLIVTPLAALLQWKSELDKFVA 123
L++ PL+ + W++E DK+ A
Sbjct 419 ------LVIVPLSTVPNWQNEFDKWAA 439
> bbo:BBOV_II004920 18.m06409; SNF2 domain-containing protein
/ helicase domain-containing protein; K10841 DNA excision repair
protein ERCC-6
Length=829
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 36/105 (34%), Positives = 54/105 (51%), Gaps = 7/105 (6%)
Query 29 VASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIP 88
V P + L Q++G+ WL + + GGILADEMG+GKT+ ++S L S I
Sbjct 138 VFCPADVFEKLYTHQKKGVKWLAEIYRNR-HGGILADEMGLGKTVTVLSFLNS----LIF 192
Query 89 ESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
+ + LIV P+ + QWK+E+ K+ L LI+H
Sbjct 193 SAEAKTLNITELKVLIVCPITLISQWKNEMIKWCP--ELKPLIFH 235
> mmu:68058 Chd1l, 4432404A22Rik, Alc1, Snf2p; chromodomain helicase
DNA binding protein 1-like (EC:3.6.4.12)
Length=900
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 44/117 (37%), Positives = 63/117 (53%), Gaps = 20/117 (17%)
Query 25 IKGRVASPE----GLL-LPLLPFQEEGLWWLTQQEASHIKGG-ILADEMGMGKTIQIISL 78
++ RV P+ GL + L +Q EG+ WL Q H + G IL DEMG+GKT Q I+L
Sbjct 21 LRTRVQEPDLQQWGLTGIRLRSYQLEGVNWLVQ--CFHCQNGCILGDEMGLGKTCQTIAL 78
Query 79 LLSRPLPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
L+ + + + P LV + PL+ L WK E+++F APG LS + Y D
Sbjct 79 LIY-LVGRLNDEGPFLV---------LCPLSVLSNWKEEMERF-APG-LSCVTYTGD 123
> bbo:BBOV_III002770 17.m07263; SNF2 family N-terminal domain
containing protein
Length=860
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 53/99 (53%), Gaps = 7/99 (7%)
Query 42 FQEEGLWWLTQQEASHIKGGILADEMGMGKTIQ---IISLLLSRPLPTIPESAPPLVR-- 96
+Q+ G+ WL+ + GILADEMG+GKT Q +S L S +P+ + ++
Sbjct 177 YQQCGVHWLSLIHSVESANGILADEMGLGKTAQASVFLSYLYSMQMPSTSKDFLSKMKLE 236
Query 97 --ACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
C STLI+ PL+ L W +EL ++ + ++ YH
Sbjct 237 AGGCRRSTLILVPLSVLDNWCNELKRWAPNLKDRIVKYH 275
> bbo:BBOV_IV011770 23.m06400; snf2-related chromatin remodeling
factor SRCAP
Length=1675
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 39/102 (38%), Positives = 50/102 (49%), Gaps = 21/102 (20%)
Query 39 LLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRAC 98
L P+Q +GL WL + GILADEMG+GKT+Q I+LL AC
Sbjct 673 LRPYQLDGLRWLASLYRNK-SNGILADEMGLGKTLQTIALLAH--------------LAC 717
Query 99 VCST----LIVTPLAALLQWKSELDKFVAPGRLSVLIYHRDP 136
LIV P + LL W+ E KF PG ++L Y+ P
Sbjct 718 DHGNWGPHLIVVPTSVLLNWEMEFKKF-CPG-FTILSYYGTP 757
> sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin
remodeling complex; required for expression of early meiotic
genes; essential helicase-related protein homologous to Snf2p
(EC:3.6.1.-); K11786 ATP-dependent helicase STH1/SNF2 [EC:3.6.4.-]
Length=1359
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 36/98 (36%), Positives = 52/98 (53%), Gaps = 13/98 (13%)
Query 39 LLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRAC 98
L +Q GL W+ +H+ G ILADEMG+GKTIQ ISL+ L + + P
Sbjct 470 LKEYQLRGLEWMVSLYNNHLNG-ILADEMGLGKTIQSISLITY--LYEVKKDIGPF---- 522
Query 99 VCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRDP 136
L++ PL+ + W E +K+ AP L+ +IY P
Sbjct 523 ----LVIVPLSTITNWTLEFEKW-APS-LNTIIYKGTP 554
> mmu:623474 Rad54b, E130016E03Rik, Fsbp, MGC67261; RAD54 homolog
B (S. cerevisiae); K10877 DNA repair and recombination protein
RAD54B [EC:3.6.4.-]
Length=886
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 34/101 (33%), Positives = 54/101 (53%), Gaps = 9/101 (8%)
Query 39 LLPFQEEGLWWLTQ----QEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPL 94
L P Q++G+ +L + A G ILADEMG+GKT+Q ISL+ + P P+
Sbjct 274 LRPHQKDGIIFLYECVMGMRAVGKCGAILADEMGLGKTLQCISLIWTLQCQG-PYGGKPV 332
Query 95 VRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
++ TLIVTP + + W+ E K++ R+ + +D
Sbjct 333 IK----KTLIVTPGSLVNNWRKEFQKWLGSERIKIFTVDQD 369
> cel:Y113G7B.14 hypothetical protein
Length=540
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 56/119 (47%), Gaps = 8/119 (6%)
Query 22 KREIKG---RVASPEGLL--LPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQII 76
K++IK + A+P G + L+P QE W+ +EA GGIL G GKT +I
Sbjct 22 KKDIKKEPCQDATPNGFVADFRLMPHQEAARDWMIDREAQEPSGGILGLAHGQGKTAIVI 81
Query 77 SLLLSRPLPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
+L+L + + ++ TLI+ P + QW E + LSV +Y+ D
Sbjct 82 ALILDQKIKCASNDKKFEQKS---PTLIIVPKRIIYQWYDEFKDRLEENALSVYLYYDD 137
> hsa:9557 CHD1L, ALC1, CHDL, FLJ22530; chromodomain helicase
DNA binding protein 1-like (EC:3.6.4.12)
Length=897
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 52/100 (52%), Gaps = 15/100 (15%)
Query 37 LPLLPFQEEGLWWLTQQEASHIKGG-ILADEMGMGKTIQIISLLLSRPLPTIPESAPPLV 95
+ L +Q EG+ WL Q+ H + G IL DEMG+GKT Q I+L + L P
Sbjct 44 IHLRSYQLEGVNWLAQR--FHCQNGCILGDEMGLGKTCQTIALFIY--LAGRLNDEGPF- 98
Query 96 RACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
LI+ PL+ L WK E+ +F APG LS + Y D
Sbjct 99 -------LILCPLSVLSNWKEEMQRF-APG-LSCVTYAGD 129
> ath:AT2G18760 CHR8; CHR8 (chromatin remodeling 8); ATP binding
/ DNA binding / helicase/ nucleic acid binding; K10841 DNA
excision repair protein ERCC-6
Length=1187
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 31/107 (28%), Positives = 53/107 (49%), Gaps = 12/107 (11%)
Query 24 EIKGRVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRP 83
+++G + PE + L +Q G+ WL + GGI+ DEMG+GKTIQ++S L S
Sbjct 370 QLEGGLNIPECIFRKLFDYQRVGVQWLWELHCQR-AGGIIGDEMGLGKTIQVLSFLGSLH 428
Query 84 LPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVL 130
+ + ++I+ P+ L QW+ E K+ + +L
Sbjct 429 FSKMYK-----------PSIIICPVTLLRQWRREAQKWYPDFHVEIL 464
> pfa:PFB0730w DEAD/DEAH box helicase, putative; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1997
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 38/109 (34%), Positives = 54/109 (49%), Gaps = 13/109 (11%)
Query 25 IKGRVASPEGLLL-PLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRP 83
+K +V P L+ L+ +Q EGL WL +++ G ILADEMG+GKTIQ ISL
Sbjct 869 VKEKVKQPSILIGGELMKYQLEGLEWLVSLYNNNLHG-ILADEMGLGKTIQTISLF---- 923
Query 84 LPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIY 132
LI+ PL+ L W SE +++ L+V+ Y
Sbjct 924 -----AYLKEFKNNINVKNLIIVPLSTLPNWISEFNRWCP--SLNVITY 965
> cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4);
K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1474
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 36/96 (37%), Positives = 56/96 (58%), Gaps = 13/96 (13%)
Query 37 LPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVR 96
L L P+Q +GL W+ +++ G ILADEMG+GKTIQ ISL+ L + ++ P
Sbjct 531 LLLKPYQIKGLEWMVSLYNNNLNG-ILADEMGLGKTIQTISLVTY--LMEVKQNNGPY-- 585
Query 97 ACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIY 132
L++ PL+ L W++E K+ AP ++ +IY
Sbjct 586 ------LVIVPLSTLSNWQNEFAKW-APS-VTTIIY 613
> tgo:TGME49_091090 SNF2 family N-terminal domain-containing protein
(EC:2.7.11.1)
Length=1345
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 63/119 (52%), Gaps = 14/119 (11%)
Query 8 ATTEAAEGMLSGLTKR---EIKGRVASPEGLLLPLLPFQEEGLWWLTQQEASHIKGGILA 64
++ E + + SGL +R EI+G L +QE+G+ WL + + G I+A
Sbjct 35 SSVERRQQVESGLAERRRKEIRGLQGVQNSAGNVLHSYQEDGVLWLIDHYLNGV-GAIVA 93
Query 65 DEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVA 123
DEMG+GKT+Q ++ + L + ++ P +L+V PL+ + W+++L F+A
Sbjct 94 DEMGLGKTLQSLAFVCW--LNEVRKAGRP--------SLVVCPLSVVSSWEAQLRDFIA 142
> sce:YFR038W IRC5; Irc5p (EC:3.6.1.-)
Length=853
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 40/95 (42%), Positives = 51/95 (53%), Gaps = 14/95 (14%)
Query 39 LLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRAC 98
L P+Q EGL WL + + G ILADEMG+GKT+Q I+LL + + P LV A
Sbjct 222 LKPYQLEGLNWLITLYENGLNG-ILADEMGLGKTVQSIALLAF--IYEMDTKGPFLVTA- 277
Query 99 VCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
PL+ L W +E KF AP L VL Y+
Sbjct 278 --------PLSTLDNWMNEFAKF-APD-LPVLKYY 302
> dre:393283 chd1l, MGC56084, zgc:56084; chromodomain helicase
DNA binding protein 1-like (EC:3.6.4.12)
Length=1026
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 55/99 (55%), Gaps = 13/99 (13%)
Query 37 LPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVR 96
+ L P+Q +G+ WL+ + +G IL DEMG+GKT Q IS LL+ ++ + P LV
Sbjct 33 IHLRPYQLDGVKWLSLCMKNQ-QGCILGDEMGLGKTCQTIS-LLAYARGSLKMNGPFLV- 89
Query 97 ACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
+C PLA L W+ EL++F LSV+ Y D
Sbjct 90 --LC------PLAVLENWRQELERFCPS--LSVICYTGD 118
> ath:AT1G50410 SNF2 domain-containing protein / helicase domain-containing
protein / RING finger domain-containing protein
Length=981
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 27/60 (45%), Positives = 39/60 (65%), Gaps = 3/60 (5%)
Query 26 KGRVASPEGLL-LPLLPFQEEGLWWLTQQEAS--HIKGGILADEMGMGKTIQIISLLLSR 82
K V P GLL +PL+ Q+ L W+ Q+E + H GGILAD+ G+GKT+ I+L+L +
Sbjct 212 KSEVDLPAGLLSVPLMKHQKIALAWMFQKETNSLHCMGGILADDQGLGKTVSTIALILKQ 271
Score = 35.8 bits (81), Expect = 0.037, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 24/41 (58%), Gaps = 6/41 (14%)
Query 102 TLIVTPLAALLQWKSELD-KFVAPGRLSVLIYH-----RDP 136
TLIV P + + QW ELD K +LSVLIYH +DP
Sbjct 345 TLIVCPASVVRQWARELDEKVTDEAKLSVLIYHGGNRTKDP 385
> tgo:TGME49_036970 SNF2 family N-terminal domain containing protein
(EC:3.6.3.8 3.6.3.14)
Length=2556
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 44/86 (51%), Gaps = 13/86 (15%)
Query 37 LPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVR 96
+ L P QEEG+ WL + + GGILADEMG+GKTIQ + L L + P
Sbjct 437 IALKPHQEEGVEWLLR--SFMTGGGILADEMGLGKTIQTLCFLSY--LNAMKLEGP---- 488
Query 97 ACVCSTLIVTPLAALLQWKSELDKFV 122
LIV PL+ + W E+ +F
Sbjct 489 -----HLIVVPLSTVGNWMREVHRFT 509
> dre:553328 hells, cb65, im:6911667, pasg, sb:cb65, sb:cb749;
helicase, lymphoid-specific
Length=853
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 35/92 (38%), Positives = 49/92 (53%), Gaps = 14/92 (15%)
Query 42 FQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCS 101
+Q EG+ WL + I G ILADEMG+GKTIQ I+ + A + + +
Sbjct 236 YQVEGIEWLRMLWENGING-ILADEMGLGKTIQCIAHI-----------AMMVEKKVLGP 283
Query 102 TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
L+V PL+ L W SE +F +SVL+YH
Sbjct 284 FLVVAPLSTLPNWISEFKRFTP--EVSVLLYH 313
> ath:AT2G28290 SYD; SYD (SPLAYED); ATPase/ chromatin binding
Length=3543
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 52/94 (55%), Gaps = 13/94 (13%)
Query 39 LLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRAC 98
L +Q GL WL +H+ GILADEMG+GKT+Q+ISL+ + T + P
Sbjct 754 LREYQMNGLRWLVSLYNNHL-NGILADEMGLGKTVQVISLICYL-METKNDRGP------ 805
Query 99 VCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIY 132
L+V P + L W+SE++ F AP + ++Y
Sbjct 806 ---FLVVVPSSVLPGWQSEIN-FWAPS-IHKIVY 834
> ath:AT2G25170 PKL; PKL (PICKLE); ATPase/ DNA binding / DNA helicase;
K11643 chromodomain-helicase-DNA-binding protein 4
[EC:3.6.4.12]
Length=1384
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 38/104 (36%), Positives = 58/104 (55%), Gaps = 19/104 (18%)
Query 31 SPEGLLLPLLPFQEEGLWWL--TQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIP 88
+PE L L P+Q EGL +L + + +H+ ILADEMG+GKTIQ I+LL S
Sbjct 265 TPEFLKGLLHPYQLEGLNFLRFSWSKQTHV---ILADEMGLGKTIQSIALLAS------- 314
Query 89 ESAPPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIY 132
L + L++ PL+ L W+ E + AP +++V++Y
Sbjct 315 -----LFEENLIPHLVIAPLSTLRNWEREFATW-AP-QMNVVMY 351
> cpv:cgd5_2920 hypothetical protein
Length=677
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/94 (32%), Positives = 50/94 (53%), Gaps = 16/94 (17%)
Query 41 PFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVC 100
P Q +G+ W+ + +GGILADEMG+GKTIQ+ + + L R+ +
Sbjct 176 PHQYDGVRWMWNR-FRRSEGGILADEMGLGKTIQVCVFIGA------------LYRSEIA 222
Query 101 S-TLIVTPLAALLQWKSELDKFVAPGRLSVLIYH 133
+ ++ P + + QWK ELDK+ ++ IYH
Sbjct 223 TFIFLILPTSLISQWKEELDKWCP--KIPKFIYH 254
> sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit
of the SWI/SNF chromatin remodeling complex involved in transcriptional
regulation; contains DNA-stimulated ATPase activity;
functions interdependently in transcriptional activation
with Snf5p and Snf6p (EC:3.6.1.-); K11786 ATP-dependent helicase
STH1/SNF2 [EC:3.6.4.-]
Length=1703
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 45/83 (54%), Gaps = 12/83 (14%)
Query 42 FQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESAPPLVRACVCS 101
+Q +GL W+ +H+ G ILADEMG+GKTIQ ISLL L + P
Sbjct 770 YQIKGLQWMVSLFNNHLNG-ILADEMGLGKTIQTISLLTY--LYEMKNIRGPY------- 819
Query 102 TLIVTPLAALLQWKSELDKFVAP 124
L++ PL+ L W SE K+ AP
Sbjct 820 -LVIVPLSTLSNWSSEFAKW-AP 840
> bbo:BBOV_IV008380 23.m05834; SNF2 helicase (EC:3.6.1.-)
Length=894
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 35/104 (33%), Positives = 54/104 (51%), Gaps = 11/104 (10%)
Query 32 PEGLLLPLLPFQEEGLWWLTQQEASHIKGGILADEMGMGKTIQIISLLLSRPLPTIPESA 91
P+ L+ P+Q EGL WL ++ G ILADEMG+GKT Q ISL L + ES
Sbjct 79 PKNLVGTAKPYQLEGLRWLVGLYDRNMNG-ILADEMGLGKTFQTISL-----LAYLKES- 131
Query 92 PPLVRACVCSTLIVTPLAALLQWKSELDKFVAPGRLSVLIYHRD 135
R L++ P + + W +E+++F R+ I +++
Sbjct 132 ----RGIDGLHLVIAPKSTIGNWINEINRFCPDLRVLKFIGNKE 171
Lambda K H
0.318 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2362384368
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40