bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
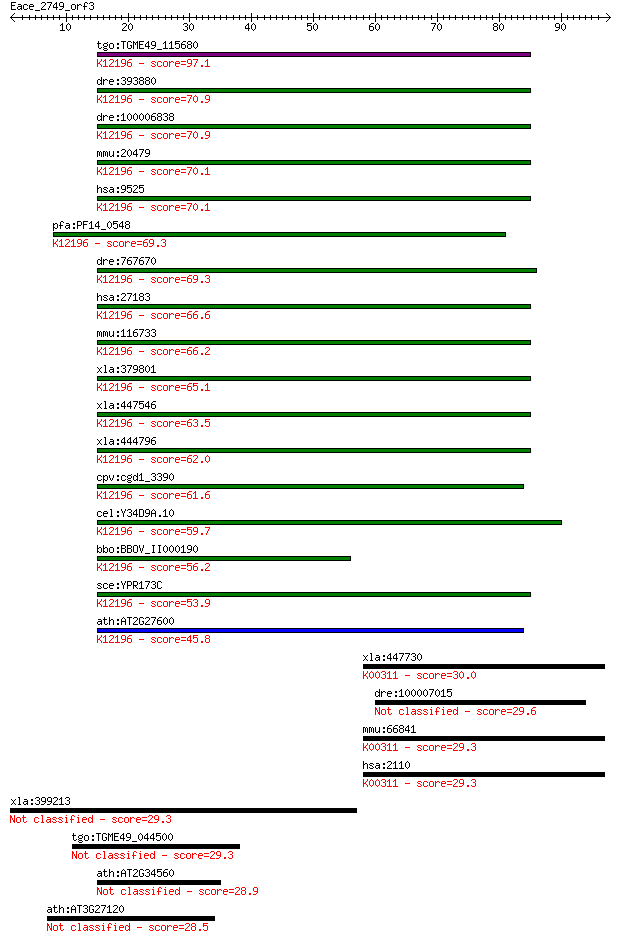
Query= Eace_2749_orf3
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12... 97.1 1e-20
dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sortin... 70.9 1e-12
dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog ... 70.9 1e-12
mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting... 70.1 2e-12
hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting ... 70.1 2e-12
pfa:PF14_0548 ATPase, putative; K12196 vacuolar protein-sortin... 69.3 3e-12
dre:767670 vps4a, MGC153907, zgc:153907; vacuolar protein sort... 69.3 3e-12
hsa:27183 VPS4A, FLJ22197, SKD1, SKD1A, SKD2, VPS4, VPS4-1; va... 66.6 2e-11
mmu:116733 Vps4a, 4930589C15Rik, AI325971, AW553189, MGC11698;... 66.2 3e-11
xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog... 65.1 5e-11
xla:447546 vps4a, vsp4; vacuolar protein sorting 4 homolog A; ... 63.5 2e-10
xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-a... 62.0 4e-10
cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196 ... 61.6 6e-10
cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting... 59.7 2e-09
bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuol... 56.2 2e-08
sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-A... 53.9 1e-07
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 45.8 3e-05
xla:447730 etfdh, MGC81928; electron-transferring-flavoprotein... 30.0 1.9
dre:100007015 hypothetical LOC100007015 29.6 2.2
mmu:66841 Etfdh, 0610010I20Rik, AV001013; electron transferrin... 29.3 3.1
hsa:2110 ETFDH, ETFQO, MADD; electron-transferring-flavoprotei... 29.3 3.1
xla:399213 lsf, Tcfcp2; transcription factor LSF 29.3 3.3
tgo:TGME49_044500 tubulin-tyrosine ligase family protein (EC:6... 29.3 3.5
ath:AT2G34560 katanin, putative 28.9 4.1
ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/... 28.5 5.5
> tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12196
vacuolar protein-sorting-associated protein 4
Length=502
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 44/70 (62%), Positives = 55/70 (78%), Gaps = 0/70 (0%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRAFYVPCRPDDSDPTTRRMTLMSVPS 74
FSG+DISV+VRDALF+P+R+CR AT FK+V +DG F PC P DSDP+ M LM VP
Sbjct 399 FSGADISVVVRDALFQPLRKCRAATHFKRVFLDGTHFLSPCPPGDSDPSKVEMRLMEVPP 458
Query 75 DRLLPPEVTM 84
+RLLPPE++M
Sbjct 459 NRLLPPELSM 468
> dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 48/78 (61%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FKQV R+ PC P DP +
Sbjct 329 YSGADISIIVRDALMQPVRKVQSATHFKQVRGPSRSDPNVIVDDLLTPCSP--GDPQAKE 386
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP ++LL P V+M
Sbjct 387 MTWMEVPGEKLLEPIVSM 404
> dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog
b-like; K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 48/78 (61%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FKQV R+ PC P DP +
Sbjct 329 YSGADISIIVRDALMQPVRKVQSATHFKQVRGPSRSDPNVIVDDLLTPCSP--GDPQAKE 386
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP ++LL P V+M
Sbjct 387 MTWMEVPGEKLLEPIVSM 404
> mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=444
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 35/78 (44%), Positives = 47/78 (60%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FK+V RA PC P DP
Sbjct 336 YSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRADPNCIVNDLLTPCSP--GDPGAIE 393
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP D+LL P V+M
Sbjct 394 MTWMDVPGDKLLEPVVSM 411
> hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting
4 homolog B (S. cerevisiae); K12196 vacuolar protein-sorting-associated
protein 4
Length=444
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 35/78 (44%), Positives = 47/78 (60%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FK+V RA PC P DP
Sbjct 336 YSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRADPNHLVDDLLTPCSP--GDPGAIE 393
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP D+LL P V+M
Sbjct 394 MTWMDVPGDKLLEPVVSM 411
> pfa:PF14_0548 ATPase, putative; K12196 vacuolar protein-sorting-associated
protein 4
Length=419
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 30/74 (40%), Positives = 46/74 (62%), Gaps = 1/74 (1%)
Query 8 FCSLWFRFSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRAFYVPCRPDDSDPTTRRM 67
F +L ++G+DI +L RDA++ P+++C + FKQV + + Y PC P DSDPT
Sbjct 309 FATLTENYTGADIDILCRDAVYMPVKKCLLSKFFKQVKKNNKICYTPCSPGDSDPTKVEK 368
Query 68 TLMSVPSDRL-LPP 80
+MS+ + L LPP
Sbjct 369 NVMSLSENELSLPP 382
> dre:767670 vps4a, MGC153907, zgc:153907; vacuolar protein sorting
4a (yeast); K12196 vacuolar protein-sorting-associated
protein 4
Length=440
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 47/79 (59%), Gaps = 10/79 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FK+V R+ PC P DP
Sbjct 328 YSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRSNSAVIVDDLLTPCSP--GDPEAIE 385
Query 67 MTLMSVPSDRLLPPEVTMV 85
MT M VP D+LL P V MV
Sbjct 386 MTWMDVPGDKLLEPIVCMV 404
> hsa:27183 VPS4A, FLJ22197, SKD1, SKD1A, SKD2, VPS4, VPS4-1;
vacuolar protein sorting 4 homolog A (S. cerevisiae); K12196
vacuolar protein-sorting-associated protein 4
Length=437
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 33/78 (42%), Positives = 45/78 (57%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRD+L +P+R+ ++AT FK+V R PC P DP
Sbjct 329 YSGADISIIVRDSLMQPVRKVQSATHFKKVCGPSRTNPSMMIDDLLTPCSP--GDPGAME 386
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP D+LL P V M
Sbjct 387 MTWMDVPGDKLLEPVVCM 404
> mmu:116733 Vps4a, 4930589C15Rik, AI325971, AW553189, MGC11698;
vacuolar protein sorting 4a (yeast); K12196 vacuolar protein-sorting-associated
protein 4
Length=437
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/78 (42%), Positives = 45/78 (57%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRD+L +P+R+ ++AT FK+V R PC P DP
Sbjct 329 YSGADISIIVRDSLMQPVRKVQSATHFKKVCGPSRTNPSVMIDDLLTPCSP--GDPGAIE 386
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP D+LL P V M
Sbjct 387 MTWMDVPGDKLLEPVVCM 404
> xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog
B; K12196 vacuolar protein-sorting-associated protein 4
Length=442
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 33/78 (42%), Positives = 44/78 (56%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVM--------VDGRAFYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FK+V V PC P DP
Sbjct 334 YSGADISIIVRDALMQPVRKVQSATHFKRVKGKSPLDPNVTRDDLLTPCSP--GDPNAVE 391
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M VP D+L P V M
Sbjct 392 MTWMDVPGDKLFEPVVCM 409
> xla:447546 vps4a, vsp4; vacuolar protein sorting 4 homolog A;
K12196 vacuolar protein-sorting-associated protein 4
Length=436
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 33/78 (42%), Positives = 44/78 (56%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRA--------FYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FK+V R PC P DP
Sbjct 328 YSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRTNPGIIVDDLLTPCSP--GDPGAVE 385
Query 67 MTLMSVPSDRLLPPEVTM 84
MT M V SD+L P V M
Sbjct 386 MTWMEVSSDKLQEPVVCM 403
> xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-associated
protein 4
Length=443
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 32/78 (41%), Positives = 44/78 (56%), Gaps = 10/78 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVM--------VDGRAFYVPCRPDDSDPTTRR 66
+SG+DIS++VRDAL +P+R+ ++AT FK+ V PC P DP
Sbjct 335 YSGADISIIVRDALMQPVRKVQSATHFKKERGKSPLDPNVTRDDLLTPCSP--GDPNAVE 392
Query 67 MTLMSVPSDRLLPPEVTM 84
MT + VP D+LL P V M
Sbjct 393 MTWVDVPGDKLLEPVVCM 410
> cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196
vacuolar protein-sorting-associated protein 4
Length=462
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 53/87 (60%), Gaps = 18/87 (20%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMV-------------DGRAFYVPC-RPDDS 60
+S SD+S+L++DALFEPIR+C + FK+V++ + + ++ PC +P +
Sbjct 341 YSSSDVSILIKDALFEPIRKCSESNWFKKVVIMNNNDEITNNNAENFKIYWTPCSQPSNI 400
Query 61 D----PTTRRMTLMSVPSDRLLPPEVT 83
D R+ +L +P+++LLPP++T
Sbjct 401 DHYDKELYRKTSLYDIPNNQLLPPKLT 427
> cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting
factor family member (vps-4); K12196 vacuolar protein-sorting-associated
protein 4
Length=430
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/83 (38%), Positives = 43/83 (51%), Gaps = 10/83 (12%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVM--------VDGRAFYVPCRPDDSDPTTRR 66
+SG DIS+LV+DAL +P+RR ++AT FK V V PC P DP
Sbjct 320 YSGYDISILVKDALMQPVRRVQSATHFKHVSGPSPKDPNVIAHDLLTPCSP--GDPHAIA 377
Query 67 MTLMSVPSDRLLPPEVTMVGAKR 89
M + VP D+L P ++M R
Sbjct 378 MNWLDVPGDKLANPPLSMQDISR 400
> bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuolar
protein-sorting-associated protein 4
Length=363
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 22/41 (53%), Positives = 33/41 (80%), Gaps = 0/41 (0%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRAFYVPC 55
+SGSD++V+VRDA +P+R+CR A+ FK+V+ +G FY PC
Sbjct 317 YSGSDVNVVVRDARMQPLRKCRDASFFKKVIRNGEEFYTPC 357
> sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-ATPase
involved in multivesicular body (MVB) protein sorting,
ATP-bound Vps4p localizes to endosomes and catalyzes ESCRT-III
disassembly and membrane release; ATPase activity is activated
by Vta1p; regulates cellular sterol metabolism; K12196
vacuolar protein-sorting-associated protein 4
Length=437
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 42/72 (58%), Gaps = 4/72 (5%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMV--DGRAFYVPCRPDDSDPTTRRMTLMSV 72
+SGSDI+V+V+DAL +PIR+ ++AT FK V D PC P D M+ +
Sbjct 334 YSGSDIAVVVKDALMQPIRKIQSATHFKDVSTEDDETRKLTPCSPGDDGAI--EMSWTDI 391
Query 73 PSDRLLPPEVTM 84
+D L P++T+
Sbjct 392 EADELKEPDLTI 403
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 41/71 (57%), Gaps = 5/71 (7%)
Query 15 FSGSDISVLVRDALFEPIRRCRTATAFKQVMVDGRAFYVPCRPDDSDPTTRRMTLMSVP- 73
FSGSD+SV V+D LFEP+R+ + A F + DG ++PC P M ++
Sbjct 334 FSGSDVSVCVKDVLFEPVRKTQDAMFFFK-SPDGT--WMPCGPRHPGAIQTTMQDLATKG 390
Query 74 -SDRLLPPEVT 83
+++++PP +T
Sbjct 391 LAEKIIPPPIT 401
> xla:447730 etfdh, MGC81928; electron-transferring-flavoprotein
dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=616
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 19/52 (36%), Gaps = 13/52 (25%)
Query 58 DDSDPTTRRMTLMSVPSDRLLPPEV-------------TMVGAKRCTYCKLC 96
DDS P R + + P R P V + A+ C +CK C
Sbjct 540 DDSTPVNRNLAIFDGPEQRFCPAGVYEYVPLESGEGSRLQINAQNCVHCKTC 591
> dre:100007015 hypothetical LOC100007015
Length=1099
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 60 SDPTTRRMTLMSVPSDRLLPPEVTMVGAKRCTYC 93
SDP+ + + +MS PSD + P V V T C
Sbjct 53 SDPSVKMLDMMSKPSDEVAKPSVMNVSTPVHTVC 86
> mmu:66841 Etfdh, 0610010I20Rik, AV001013; electron transferring
flavoprotein, dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=616
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 20/52 (38%), Gaps = 13/52 (25%)
Query 58 DDSDPTTRRMTLMSVPSDRLLPPEV-------------TMVGAKRCTYCKLC 96
DDS P R +++ P R P V + A+ C +CK C
Sbjct 540 DDSIPVNRNLSIYDGPEQRFCPAGVYEFVPLEQGDGFRLQINAQNCVHCKTC 591
> hsa:2110 ETFDH, ETFQO, MADD; electron-transferring-flavoprotein
dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=617
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 20/52 (38%), Gaps = 13/52 (25%)
Query 58 DDSDPTTRRMTLMSVPSDRLLPPEV-------------TMVGAKRCTYCKLC 96
DDS P R +++ P R P V + A+ C +CK C
Sbjct 541 DDSIPVNRNLSIYDGPEQRFCPAGVYEFVPVEQGDGFRLQINAQNCVHCKTC 592
> xla:399213 lsf, Tcfcp2; transcription factor LSF
Length=506
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 26/59 (44%), Gaps = 3/59 (5%)
Query 1 YSTRFLLFCSLWFRFSGSDISVLVRDALFE---PIRRCRTATAFKQVMVDGRAFYVPCR 56
+ RF F L+ FSG+D+ L RD + + P R A K MV R C+
Sbjct 347 HRNRFAAFSRLFTNFSGADLLKLTRDDIIQICGPADGIRLFNALKGRMVRPRLTVYVCQ 405
> tgo:TGME49_044500 tubulin-tyrosine ligase family protein (EC:6.3.2.25)
Length=2387
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 14/27 (51%), Gaps = 0/27 (0%)
Query 11 LWFRFSGSDISVLVRDALFEPIRRCRT 37
WF FS I V R AL P R+C T
Sbjct 1508 FWFLFSPQTIQVSSRAALQSPARKCHT 1534
> ath:AT2G34560 katanin, putative
Length=384
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 10/20 (50%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 15 FSGSDISVLVRDALFEPIRR 34
+SGSDI +L ++A +P+RR
Sbjct 306 YSGSDIRILCKEAAMQPLRR 325
> ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=476
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 7 LFCSLWFRFSGSDISVLVRDALFEPIR 33
+ C+L +SGSD+ LV+DA P+R
Sbjct 394 IICNLTEGYSGSDMKNLVKDATMGPLR 420
Lambda K H
0.332 0.142 0.467
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40