bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2750_orf1
Length=83
Score E
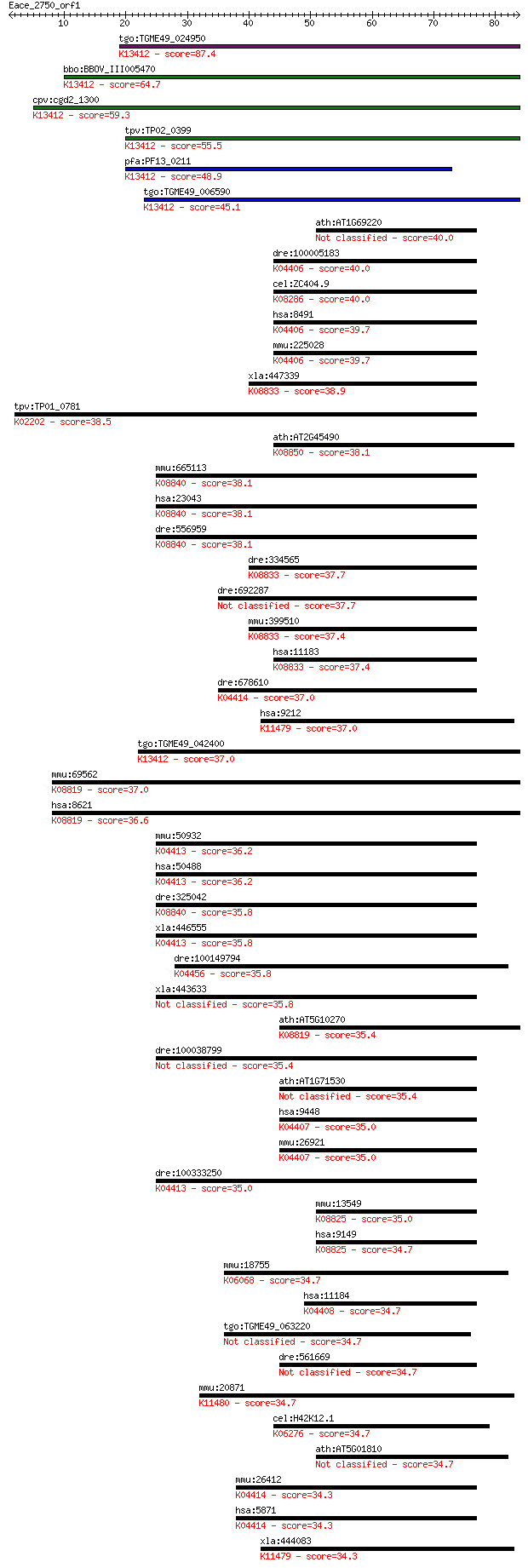
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 87.4 1e-17
bbo:BBOV_III005470 17.m07489; protein kinase domain containing... 64.7 7e-11
cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with... 59.3 3e-09
tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium... 55.5 4e-08
pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2... 48.9 4e-06
tgo:TGME49_006590 calcium-dependent protein kinase, putative (... 45.1 5e-05
ath:AT1G69220 SIK1; SIK1; ATP binding / kinase/ protein kinase... 40.0 0.002
dre:100005183 map4k3; mitogen-activated protein kinase kinase ... 40.0 0.002
cel:ZC404.9 gck-2; Germinal Center Kinase family member (gck-2... 40.0 0.002
hsa:8491 MAP4K3, GLK, MAPKKKK3, MEKKK_3, RAB8IPL1; mitogen-act... 39.7 0.002
mmu:225028 Map4k3, 4833416M01Rik, 4833416M07Rik, 9530052P13Rik... 39.7 0.002
xla:447339 map4k5, MGC83247; mitogen-activated protein kinase ... 38.9 0.004
tpv:TP01_0781 cyclin-dependent kinase; K02202 cyclin-dependent... 38.5 0.006
ath:AT2G45490 AtAUR3; AtAUR3 (ATAURORA3); ATP binding / histon... 38.1 0.006
mmu:665113 Tnik, 1500031A17Rik, 4831440I19Rik, AI451411, C5300... 38.1 0.006
hsa:23043 TNIK; TRAF2 and NCK interacting kinase (EC:2.7.11.1)... 38.1 0.007
dre:556959 tnikb, MGC123033, MGC152783, MGC192776, im:6898857,... 38.1 0.007
dre:334565 map4k5, MGC55719, fl74c10, wu:fl74c10, zgc:55719, z... 37.7 0.009
dre:692287 zgc:136354 (EC:2.7.11.1) 37.7 0.009
mmu:399510 Map4k5, 4432415E19Rik, GCKR, KHS, MAPKKKK5; mitogen... 37.4 0.011
hsa:11183 MAP4K5, GCKR, KHS, KHS1, MAPKKKK5; mitogen-activated... 37.4 0.012
dre:678610 map4k2l, MGC136670, dZ150L22.2, map4k3l, si:dz150l2... 37.0 0.014
hsa:9212 AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, STK12, ST... 37.0 0.014
tgo:TGME49_042400 calcium-dependent protein kinase, putative (... 37.0 0.017
mmu:69562 Cdk13, 2310015O17Rik, Cdc2l5; cyclin-dependent kinas... 37.0 0.017
hsa:8621 CDK13, CDC2L, CDC2L5, CHED, FLJ35215, KIAA1791; cycli... 36.6 0.019
mmu:50932 Mink1, B55, MINK, Map4k6, RP23-122P1.6, Ysk2; missha... 36.2 0.029
hsa:50488 MINK1, B55, MAP4K6, MGC21111, MINK, YSK2, ZC3, hMINK... 36.2 0.029
dre:325042 tnika, fc52d11, msn, tnikl, wu:fc52d11, zgc:63535; ... 35.8 0.031
xla:446555 mink1, MGC81192, mink; misshapen-like kinase 1 (EC:... 35.8 0.031
dre:100149794 akt3; v-akt murine thymoma viral oncogene homolo... 35.8 0.033
xla:443633 tnik, MGC130721; TRAF2 and NCK interacting kinase 35.8 0.035
ath:AT5G10270 CDKC;1; CDKC;1 (Cyclin-dependent kinase C;1); ki... 35.4 0.042
dre:100038799 mink1, si:ch211-147a23.2, zgc:162163; misshapen-... 35.4 0.042
ath:AT1G71530 protein kinase family protein 35.4 0.044
hsa:9448 MAP4K4, FLH21957, FLJ10410, FLJ20373, FLJ90111, HGK, ... 35.0 0.054
mmu:26921 Map4k4, 9430080K19Rik, AU043147, AU045934, AW046177,... 35.0 0.055
dre:100333250 misshapen-like kinase 1-like; K04413 misshapen/N... 35.0 0.057
mmu:13549 Dyrk1b, Mirk; dual-specificity tyrosine-(Y)-phosphor... 35.0 0.066
hsa:9149 DYRK1B, MIRK; dual-specificity tyrosine-(Y)-phosphory... 34.7 0.069
mmu:18755 Prkch, Pkch; protein kinase C, eta (EC:2.7.11.13); K... 34.7 0.071
hsa:11184 MAP4K1, HPK1; mitogen-activated protein kinase kinas... 34.7 0.071
tgo:TGME49_063220 protein kinase, putative (EC:2.7.11.17) 34.7 0.073
dre:561669 mitogen-activated protein kinase kinase kinase kina... 34.7 0.073
mmu:20871 Aurkc, AIE1, AIK3, IAK3, Stk13; aurora kinase C (EC:... 34.7 0.077
cel:H42K12.1 pdk-1; PDK-class protein kinase family member (pd... 34.7 0.078
ath:AT5G01810 CIPK15; CIPK15 (CBL-INTERACTING PROTEIN KINASE 1... 34.7 0.080
mmu:26412 Map4k2, AI385662, BL44, GCK, MGC143893, MGC143894, R... 34.3 0.088
hsa:5871 MAP4K2, BL44, GCK, RAB8IP; mitogen-activated protein ... 34.3 0.088
xla:444083 aurkb-b, AurB, MGC83575, aik2, aim1, airk2, ark2, i... 34.3 0.092
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 40/65 (61%), Positives = 49/65 (75%), Gaps = 0/65 (0%)
Query 19 GPVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYK 78
PVFDRS+ V E+ L I DI+D + +LGKGSYG VVKA+DLKTG +RAIK+VYK
Sbjct 187 APVFDRSKIVHEVLLKDGQQITDIYDTPNGTSLGKGSYGSVVKAKDLKTGIMRAIKIVYK 246
Query 79 PRIEN 83
PRI+N
Sbjct 247 PRIDN 251
> bbo:BBOV_III005470 17.m07489; protein kinase domain containing
protein; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=755
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 46/75 (61%), Gaps = 2/75 (2%)
Query 10 NTEKEVHV-AGPVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTG 68
N K+ H + P FDRS + E L +I D++D H LGKGSYGQV+KA +TG
Sbjct 265 NDPKQGHRDSTPTFDRSFLIHETALVDGMTITDVYDL-HTNRLGKGSYGQVLKACHKETG 323
Query 69 SIRAIKVVYKPRIEN 83
++A+KV+ K IEN
Sbjct 324 EVKAVKVIRKAAIEN 338
> cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=677
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 33/85 (38%), Positives = 56/85 (65%), Gaps = 8/85 (9%)
Query 5 GEQQHNTEKEVH------VAGPVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQ 58
GE++ +T+ + V+ +FDR+ +QE L + +IND ++ + NLG+GSYG
Sbjct 141 GERETSTDSNIKNTESTKVSHGIFDRTCLIQEHALV-NRNINDFYEL-NLGNLGRGSYGS 198
Query 59 VVKARDLKTGSIRAIKVVYKPRIEN 83
VVKA D ++G+ RA+K++ KP++EN
Sbjct 199 VVKAIDKQSGAQRAVKIILKPKLEN 223
> tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=844
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 20 PVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKP 79
P+ +RS+ + + L TSI+D+++ ++ LG GSYG V+K ++G++RAIK++ K
Sbjct 376 PLLNRSKIIPQTALTDGTSIHDVYEIK-SEKLGNGSYGNVLKGVHKESGAVRAIKIILKS 434
Query 80 RIEN 83
+IEN
Sbjct 435 KIEN 438
> pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=568
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 25/53 (47%), Positives = 35/53 (66%), Gaps = 1/53 (1%)
Query 20 PVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRA 72
P FDRS+ +QEI L + ++D+++ + LGKGSYG VVKA +TG RA
Sbjct 101 PYFDRSQIIQEIILMNNDELSDVYEIDRYK-LGKGSYGNVVKAVSKRTGQQRA 152
> tgo:TGME49_006590 calcium-dependent protein kinase, putative
(EC:2.7.11.17 3.4.22.53); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=761
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 39/61 (63%), Gaps = 5/61 (8%)
Query 23 DRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRIE 82
DRS+F+ L+ + ++ D ++ A LG+G+YG V KAR TG +RA+K + K +++
Sbjct 208 DRSKFI----LENTGALTDFYEIDTA-TLGQGTYGSVSKARKKDTGQMRAVKTISKSQVK 262
Query 83 N 83
N
Sbjct 263 N 263
> ath:AT1G69220 SIK1; SIK1; ATP binding / kinase/ protein kinase/
protein serine/threonine kinase
Length=836
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 19/26 (73%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 51 LGKGSYGQVVKARDLKTGSIRAIKVV 76
LGKGSYG V KARDLKT I A+KV+
Sbjct 255 LGKGSYGSVYKARDLKTSEIVAVKVI 280
> dre:100005183 map4k3; mitogen-activated protein kinase kinase
kinase kinase 3; K04406 mitogen-activated protein kinase kinase
kinase kinase 3 [EC:2.7.11.1]
Length=897
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 17/33 (51%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
DF Q +G G+YG V KAR++ TG + AIKV+
Sbjct 15 DFELIQRIGSGTYGDVYKARNVSTGELAAIKVI 47
> cel:ZC404.9 gck-2; Germinal Center Kinase family member (gck-2);
K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=829
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
D+ Q +G G+YG+V KARD+++ S+ A+KVV
Sbjct 14 DYELLQRVGSGTYGEVYKARDIRSDSLAAVKVV 46
> hsa:8491 MAP4K3, GLK, MAPKKKK3, MEKKK_3, RAB8IPL1; mitogen-activated
protein kinase kinase kinase kinase 3 (EC:2.7.11.1);
K04406 mitogen-activated protein kinase kinase kinase kinase
3 [EC:2.7.11.1]
Length=894
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 17/33 (51%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
DF Q +G G+YG V KAR++ TG + AIKV+
Sbjct 15 DFELIQRIGSGTYGDVYKARNVNTGELAAIKVI 47
> mmu:225028 Map4k3, 4833416M01Rik, 4833416M07Rik, 9530052P13Rik,
Glk, MAPKKKK3, RAB8IPL1; mitogen-activated protein kinase
kinase kinase kinase 3 (EC:2.7.11.1); K04406 mitogen-activated
protein kinase kinase kinase kinase 3 [EC:2.7.11.1]
Length=896
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 17/33 (51%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
DF Q +G G+YG V KAR++ TG + AIKV+
Sbjct 15 DFELIQRIGSGTYGDVYKARNVNTGELAAIKVI 47
> xla:447339 map4k5, MGC83247; mitogen-activated protein kinase
kinase kinase kinase 5 (EC:2.7.11.1); K08833 mitogen-activated
protein kinase kinase kinase kinase 5 [EC:2.7.11.1]
Length=823
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 40 NDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
N D+ Q +G G+YG V KAR+L+TG + A+K++
Sbjct 11 NPQHDYELIQRVGSGTYGDVYKARNLQTGELAAVKII 47
> tpv:TP01_0781 cyclin-dependent kinase; K02202 cyclin-dependent
kinase 7 [EC:2.7.11.22]
Length=415
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 41/80 (51%), Gaps = 10/80 (12%)
Query 2 LEAGEQQHNTEKEVHVAGPVFDRSRFVQEIQLDGSTSINDIFDFTH-----AQNLGKGSY 56
LE G+ N EK V V FD +++ + D T N+ ++LG+G+Y
Sbjct 46 LEDGKNVENAEKTVKVE---FDDK--IEDKKEDSVTDSNESESLDKRFTPVGKHLGEGTY 100
Query 57 GQVVKARDLKTGSIRAIKVV 76
GQV+KA D TG + AIK V
Sbjct 101 GQVIKAMDTLTGKMVAIKKV 120
> ath:AT2G45490 AtAUR3; AtAUR3 (ATAURORA3); ATP binding / histone
kinase(H3-S10 specific) / protein kinase; K08850 aurora
kinase, other [EC:2.7.11.1]
Length=288
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 18/39 (46%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRIE 82
DF + LGKG +G+V AR+ K+ I A+KV++K +IE
Sbjct 21 DFEIGRPLGKGKFGRVYLAREAKSKYIVALKVIFKEQIE 59
> mmu:665113 Tnik, 1500031A17Rik, 4831440I19Rik, AI451411, C530008O15Rik,
C630040K21Rik, KIAA0551, MGC189819, MGC189859; TRAF2
and NCK interacting kinase (EC:2.7.11.1); K08840 TRAF2
and NCK interacting kinase [EC:2.7.11.1]
Length=1352
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + EI L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDEIDLSALRDPAGIFELVEL--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> hsa:23043 TNIK; TRAF2 and NCK interacting kinase (EC:2.7.11.1);
K08840 TRAF2 and NCK interacting kinase [EC:2.7.11.1]
Length=1352
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + EI L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDEIDLSALRDPAGIFELVEL--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> dre:556959 tnikb, MGC123033, MGC152783, MGC192776, im:6898857,
zgc:123033, zgc:152783; TRAF2 and NCK interacting kinase
b; K08840 TRAF2 and NCK interacting kinase [EC:2.7.11.1]
Length=1329
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + EI L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDEIDLSALRDPAGIFELVEL--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> dre:334565 map4k5, MGC55719, fl74c10, wu:fl74c10, zgc:55719,
zgc:55985; mitogen-activated protein kinase kinase kinase kinase
5 (EC:2.7.11.1); K08833 mitogen-activated protein kinase
kinase kinase kinase 5 [EC:2.7.11.1]
Length=878
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 40 NDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
N DF Q +G G+YG V KAR + TG + A+K++
Sbjct 15 NPQHDFELIQRVGSGTYGDVYKARKISTGELAAVKII 51
> dre:692287 zgc:136354 (EC:2.7.11.1)
Length=889
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 35 GSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
G + + + D+ +G G+YG V KAR +KT I AIKVV
Sbjct 5 GVSHTDPLDDYELIHRIGSGTYGDVFKARSIKTSVIAAIKVV 46
> mmu:399510 Map4k5, 4432415E19Rik, GCKR, KHS, MAPKKKK5; mitogen-activated
protein kinase kinase kinase kinase 5 (EC:2.7.11.1);
K08833 mitogen-activated protein kinase kinase kinase
kinase 5 [EC:2.7.11.1]
Length=847
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 40 NDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
N D+ Q +G G+YG V KAR++ TG + A+K++
Sbjct 15 NPQHDYELVQRVGSGTYGDVYKARNVHTGELAAVKII 51
> hsa:11183 MAP4K5, GCKR, KHS, KHS1, MAPKKKK5; mitogen-activated
protein kinase kinase kinase kinase 5 (EC:2.7.11.1); K08833
mitogen-activated protein kinase kinase kinase kinase 5 [EC:2.7.11.1]
Length=846
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
D+ Q +G G+YG V KAR++ TG + A+K++
Sbjct 19 DYELVQRVGSGTYGDVYKARNVHTGELAAVKII 51
> dre:678610 map4k2l, MGC136670, dZ150L22.2, map4k3l, si:dz150l22.2,
si:dz263j20.1, zgc:136670; mitogen-activated protein
kinase kinase kinase kinase 2-like; K04414 mitogen-activated
protein kinase kinase kinase kinase 2 [EC:2.7.11.1]
Length=865
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 35 GSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
G + ++ + D+ Q +G G+YG V KAR++++ + AIK+V
Sbjct 5 GVSFLDPLDDYEIIQRIGSGTYGDVFKARNIRSSEMAAIKIV 46
> hsa:9212 AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, STK12,
STK5, aurkb-sv1, aurkb-sv2; aurora kinase B (EC:2.7.11.1); K11479
aurora kinase B [EC:2.7.11.1]
Length=344
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 42 IFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRIE 82
I DF + LGKG +G V AR+ K+ I A+KV++K +IE
Sbjct 74 IDDFEIGRPLGKGKFGNVYLAREKKSHFIVALKVLFKSQIE 114
> tgo:TGME49_042400 calcium-dependent protein kinase, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=604
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 35/62 (56%), Gaps = 5/62 (8%)
Query 22 FDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRI 81
F R F+ L S + D +D + LG+G+YG V +A + T ++RA+K + K ++
Sbjct 111 FRREGFI----LSYSGPLTDYYD-VETKKLGQGTYGSVCRAVNKATKNVRAVKTIPKAKV 165
Query 82 EN 83
+N
Sbjct 166 KN 167
> mmu:69562 Cdk13, 2310015O17Rik, Cdc2l5; cyclin-dependent kinase
13 (EC:2.7.11.23 2.7.11.22); K08819 Cdc2-related kinase,
arginine/serine-rich [EC:2.7.11.22]
Length=1511
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 8/76 (10%)
Query 8 QHNTEKEVHVAGPVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKT 67
Q ++++ + GP + ++E +D D FD +G+G+YGQV KARD T
Sbjct 673 QLHSKRRPKICGPRYGE---IKEKDIDWGKRCVDKFDIIGI--IGEGTYGQVYKARDKDT 727
Query 68 GSIRAIKVVYKPRIEN 83
G + A+K K R++N
Sbjct 728 GEMVALK---KVRLDN 740
> hsa:8621 CDK13, CDC2L, CDC2L5, CHED, FLJ35215, KIAA1791; cyclin-dependent
kinase 13 (EC:2.7.11.22); K08819 Cdc2-related
kinase, arginine/serine-rich [EC:2.7.11.22]
Length=1512
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 8/76 (10%)
Query 8 QHNTEKEVHVAGPVFDRSRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKT 67
Q ++++ + GP + ++ E +D D FD +G+G+YGQV KARD T
Sbjct 673 QLHSKRRPKICGPRYGETK---EKDIDWGKRCVDKFDIIGI--IGEGTYGQVYKARDKDT 727
Query 68 GSIRAIKVVYKPRIEN 83
G + A+K K R++N
Sbjct 728 GEMVALK---KVRLDN 740
> mmu:50932 Mink1, B55, MINK, Map4k6, RP23-122P1.6, Ysk2; misshapen-like
kinase 1 (zebrafish) (EC:2.7.11.1); K04413 misshapen/NIK-related
kinase [EC:2.7.11.1]
Length=1337
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + +I L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDDIDLSALRDPAGIFELVEV--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> hsa:50488 MINK1, B55, MAP4K6, MGC21111, MINK, YSK2, ZC3, hMINK,
hMINKbeta; misshapen-like kinase 1 (EC:2.7.11.1); K04413
misshapen/NIK-related kinase [EC:2.7.11.1]
Length=1312
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + +I L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDDIDLSALRDPAGIFELVEV--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> dre:325042 tnika, fc52d11, msn, tnikl, wu:fc52d11, zgc:63535;
TRAF2 and NCK interacting kinase a; K08840 TRAF2 and NCK interacting
kinase [EC:2.7.11.1]
Length=1303
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + EI L IF+ +G G+YGQV K R +KTG + AIK +
Sbjct 7 ARSLDEIDLSTLRDPAGIFELVEL--VGNGTYGQVYKGRHVKTGQLAAIKCM 56
> xla:446555 mink1, MGC81192, mink; misshapen-like kinase 1 (EC:2.7.11.1);
K04413 misshapen/NIK-related kinase [EC:2.7.11.1]
Length=1270
Score = 35.8 bits (81), Expect = 0.031, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + +I L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDDIDLSALRDPAGIFELVEV--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> dre:100149794 akt3; v-akt murine thymoma viral oncogene homolog
3 (protein kinase B, gamma); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=423
Score = 35.8 bits (81), Expect = 0.033, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 36/57 (63%), Gaps = 3/57 (5%)
Query 28 VQEIQLDGSTSIND---IFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRI 81
V E ++D STS + + DF + + LGKG++G+V+ R+ +G+ A+K++ K I
Sbjct 72 VNEEEMDTSTSHHKRKTMNDFDYLKLLGKGTFGKVILVREKASGTYYAMKILKKEVI 128
> xla:443633 tnik, MGC130721; TRAF2 and NCK interacting kinase
Length=485
Score = 35.8 bits (81), Expect = 0.035, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + EI L IF+ + +G G+YGQV K R +KTG + AIKV+
Sbjct 7 ARSLDEIDLSALRDPAGIFELV--ELVGNGTYGQVYKGRHVKTGQLAAIKVM 56
> ath:AT5G10270 CDKC;1; CDKC;1 (Cyclin-dependent kinase C;1);
kinase; K08819 Cdc2-related kinase, arginine/serine-rich [EC:2.7.11.22]
Length=505
Score = 35.4 bits (80), Expect = 0.042, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 27/39 (69%), Gaps = 3/39 (7%)
Query 45 FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRIEN 83
F + +G+G+YGQV A+++KTG I A+K K R++N
Sbjct 26 FEKLEQIGEGTYGQVYMAKEIKTGEIVALK---KIRMDN 61
> dre:100038799 mink1, si:ch211-147a23.2, zgc:162163; misshapen-like
kinase 1 (EC:2.7.11.1)
Length=150
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + +I L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 TRSLDDIDLAALRDPAGIFELVEV--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> ath:AT1G71530 protein kinase family protein
Length=463
Score = 35.4 bits (80), Expect = 0.044, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 45 FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
F +G+G+Y V KARDL+TG I A+K V
Sbjct 147 FEKLDKIGQGTYSSVYKARDLETGKIVAMKKV 178
> hsa:9448 MAP4K4, FLH21957, FLJ10410, FLJ20373, FLJ90111, HGK,
KIAA0687, NIK; mitogen-activated protein kinase kinase kinase
kinase 4 (EC:2.7.11.1); K04407 mitogen-activated protein
kinase kinase kinase kinase 4 [EC:2.7.11.1]
Length=1166
Score = 35.0 bits (79), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 45 FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
F + +G G+YGQV K R +KTG + AIKV+
Sbjct 25 FELVEVVGNGTYGQVYKGRHVKTGQLAAIKVM 56
> mmu:26921 Map4k4, 9430080K19Rik, AU043147, AU045934, AW046177,
HGK, Nik; mitogen-activated protein kinase kinase kinase
kinase 4 (EC:2.7.11.1); K04407 mitogen-activated protein kinase
kinase kinase kinase 4 [EC:2.7.11.1]
Length=1234
Score = 35.0 bits (79), Expect = 0.055, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 45 FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
F + +G G+YGQV K R +KTG + AIKV+
Sbjct 25 FELVEVVGNGTYGQVYKGRHVKTGQLAAIKVM 56
> dre:100333250 misshapen-like kinase 1-like; K04413 misshapen/NIK-related
kinase [EC:2.7.11.1]
Length=1273
Score = 35.0 bits (79), Expect = 0.057, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 25 SRFVQEIQLDGSTSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
+R + +I L IF+ +G G+YGQV K R +KTG + AIKV+
Sbjct 7 TRSLDDIDLAALRDPAGIFELVEV--VGNGTYGQVYKGRHVKTGQLAAIKVM 56
> mmu:13549 Dyrk1b, Mirk; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 1b (EC:2.7.12.1); K08825 dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase [EC:2.7.12.1]
Length=629
Score = 35.0 bits (79), Expect = 0.066, Method: Composition-based stats.
Identities = 15/26 (57%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 51 LGKGSYGQVVKARDLKTGSIRAIKVV 76
+GKGS+GQVVKA D +T + AIK++
Sbjct 117 IGKGSFGQVVKAYDHQTQELVAIKII 142
> hsa:9149 DYRK1B, MIRK; dual-specificity tyrosine-(Y)-phosphorylation
regulated kinase 1B (EC:2.7.12.1); K08825 dual-specificity
tyrosine-(Y)-phosphorylation regulated kinase [EC:2.7.12.1]
Length=629
Score = 34.7 bits (78), Expect = 0.069, Method: Composition-based stats.
Identities = 15/26 (57%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 51 LGKGSYGQVVKARDLKTGSIRAIKVV 76
+GKGS+GQVVKA D +T + AIK++
Sbjct 117 IGKGSFGQVVKAYDHQTQELVAIKII 142
> mmu:18755 Prkch, Pkch; protein kinase C, eta (EC:2.7.11.13);
K06068 novel protein kinase C [EC:2.7.11.13]
Length=683
Score = 34.7 bits (78), Expect = 0.071, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 36 STSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRI 81
S+S I +F + LGKGS+G+V+ AR +TG + A+KV+ K I
Sbjct 346 SSSRFGIDNFEFIRVLGKGSFGKVMLARIKETGELYAVKVLKKDVI 391
> hsa:11184 MAP4K1, HPK1; mitogen-activated protein kinase kinase
kinase kinase 1 (EC:2.7.11.1); K04408 mitogen-activated
protein kinase kinase kinase kinase 1 [EC:2.7.11.1]
Length=821
Score = 34.7 bits (78), Expect = 0.071, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 49 QNLGKGSYGQVVKARDLKTGSIRAIKVV 76
Q LG G+YG+V KARD +G + A+K+V
Sbjct 21 QRLGGGTYGEVFKARDKVSGDLVALKMV 48
> tgo:TGME49_063220 protein kinase, putative (EC:2.7.11.17)
Length=673
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 19/40 (47%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 36 STSINDIFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKV 75
ST N+ F LG G YG V ARDL+TG A+KV
Sbjct 324 STRTNEKFSVQLEDVLGAGGYGVVFSARDLRTGQGIALKV 363
> dre:561669 mitogen-activated protein kinase kinase kinase kinase
4-like
Length=1040
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 45 FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
F + +G G+YGQV K R +KTG + AIKV+
Sbjct 25 FELIEVVGNGTYGQVYKGRHVKTGQLAAIKVM 56
> mmu:20871 Aurkc, AIE1, AIK3, IAK3, Stk13; aurora kinase C (EC:2.7.11.1);
K11480 aurora kinase C [EC:2.7.11.1]
Length=315
Score = 34.7 bits (78), Expect = 0.077, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 32 QLDGSTSINDIF---DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRIE 82
Q++ STS F DF + LG+G +G+V AR + I A+KV++K IE
Sbjct 39 QMEPSTSTRKHFTINDFEIGRPLGRGKFGRVYLARLKENHFIVALKVLFKSEIE 92
> cel:H42K12.1 pdk-1; PDK-class protein kinase family member (pdk-1);
K06276 3-phosphoinositide dependent protein kinase-1
[EC:2.7.11.1]
Length=636
Score = 34.7 bits (78), Expect = 0.078, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 44 DFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYK 78
DF Q++G+G+Y QV + R++ T ++ A+KV+ K
Sbjct 68 DFMFLQSMGEGAYSQVFRCREVATDAMFAVKVLQK 102
> ath:AT5G01810 CIPK15; CIPK15 (CBL-INTERACTING PROTEIN KINASE
15); kinase/ protein kinase
Length=421
Score = 34.7 bits (78), Expect = 0.080, Method: Composition-based stats.
Identities = 17/31 (54%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 51 LGKGSYGQVVKARDLKTGSIRAIKVVYKPRI 81
LG+G++ +V AR LKTG AIKV+ K RI
Sbjct 18 LGQGTFAKVYHARHLKTGDSVAIKVIDKERI 48
> mmu:26412 Map4k2, AI385662, BL44, GCK, MGC143893, MGC143894,
Rab8ip; mitogen-activated protein kinase kinase kinase kinase
2 (EC:2.7.11.1); K04414 mitogen-activated protein kinase
kinase kinase kinase 2 [EC:2.7.11.1]
Length=821
Score = 34.3 bits (77), Expect = 0.088, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 38 SINDIFD-FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
S+ D D F Q +G G+YG V KARD T + A+K+V
Sbjct 8 SLQDPRDRFELLQRVGAGTYGDVYKARDTVTSELAAVKIV 47
> hsa:5871 MAP4K2, BL44, GCK, RAB8IP; mitogen-activated protein
kinase kinase kinase kinase 2 (EC:2.7.11.1); K04414 mitogen-activated
protein kinase kinase kinase kinase 2 [EC:2.7.11.1]
Length=820
Score = 34.3 bits (77), Expect = 0.088, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 38 SINDIFD-FTHAQNLGKGSYGQVVKARDLKTGSIRAIKVV 76
S+ D D F Q +G G+YG V KARD T + A+K+V
Sbjct 8 SLQDPRDRFELLQRVGAGTYGDVYKARDTVTSELAAVKIV 47
> xla:444083 aurkb-b, AurB, MGC83575, aik2, aim1, airk2, ark2,
ipl1, stk12, stk5; aurora kinase B (EC:2.7.11.1); K11479 aurora
kinase B [EC:2.7.11.1]
Length=368
Score = 34.3 bits (77), Expect = 0.092, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 42 IFDFTHAQNLGKGSYGQVVKARDLKTGSIRAIKVVYKPRIE 82
I DF + LGKG +G V AR+ + I A+KV++K ++E
Sbjct 97 IDDFDIGRPLGKGKFGNVYLAREKQNKFIMALKVLFKSQLE 137
Lambda K H
0.315 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40