bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2773_orf3
Length=93
Score E
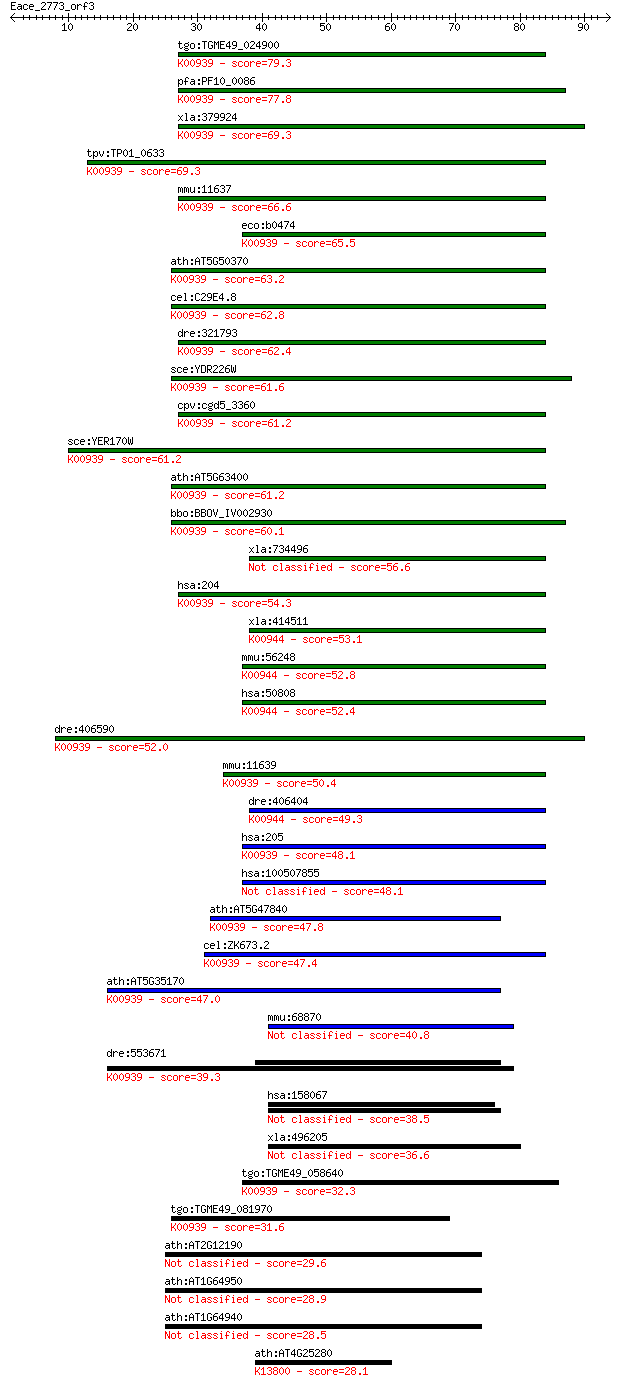
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_024900 adenylate kinase, putative (EC:2.7.4.10); K0... 79.3 3e-15
pfa:PF10_0086 adenylate kinase; K00939 adenylate kinase [EC:2.... 77.8 8e-15
xla:379924 ak2, MGC53010; adenylate kinase 2 (EC:2.7.4.3); K00... 69.3 3e-12
tpv:TP01_0633 adenylate kinase; K00939 adenylate kinase [EC:2.... 69.3 3e-12
mmu:11637 Ak2, Ak-2, D4Ertd220e; adenylate kinase 2 (EC:2.7.4.... 66.6 2e-11
eco:b0474 adk, dnaW, ECK0468, JW0463, plsA; adenylate kinase (... 65.5 4e-11
ath:AT5G50370 adenylate kinase, putative (EC:2.7.4.3); K00939 ... 63.2 2e-10
cel:C29E4.8 let-754; LEThal family member (let-754); K00939 ad... 62.8 3e-10
dre:321793 ak2, wu:fb34f05, wu:fj80e03; adenylate kinase 2 (EC... 62.4 4e-10
sce:YDR226W ADK1, AKY1, AKY2; Adenylate kinase, required for p... 61.6 6e-10
cpv:cgd5_3360 adenylate kinase ; K00939 adenylate kinase [EC:2... 61.2 7e-10
sce:YER170W ADK2, AKY3, PAK3; Adk2p (EC:2.7.4.3); K00939 adeny... 61.2 8e-10
ath:AT5G63400 ADK1; ADK1 (ADENYLATE KINASE 1); ATP binding / a... 61.2 8e-10
bbo:BBOV_IV002930 21.m02778; adenylate kinase 2 (EC:2.7.4.3); ... 60.1 2e-09
xla:734496 ak3, MGC114646, ak6; adenylate kinase 3 (EC:2.7.4.10) 56.6 2e-08
hsa:204 AK2, ADK2, AK_2; adenylate kinase 2 (EC:2.7.4.3); K009... 54.3 1e-07
xla:414511 hypothetical protein MGC82246; K00944 nucleoside-tr... 53.1 2e-07
mmu:56248 Ak3, AA407498, AI506714, AK-3, Ak3l, Ak3l1, Akl3l; a... 52.8 3e-07
hsa:50808 AK3, AK3L1, AK6, AKL3L, AKL3L1, FIX; adenylate kinas... 52.4 4e-07
dre:406590 ak4, ak3l1, wu:fc37g02, zgc:85790; adenylate kinase... 52.0 5e-07
mmu:11639 Ak4, Ak-3, Ak-4, Ak3, Ak3l1, D4Ertd274e; adenylate k... 50.4 1e-06
dre:406404 ak3, ak3l1, id:ibd3034, sb:cb845, wu:fa02c11, wu:fc... 49.3 3e-06
hsa:205 AK4, AK3, AK3L1, AK3L2, MGC166959; adenylate kinase 4 ... 48.1 7e-06
hsa:100507855 adenylate kinase isoenzyme 4, mitochondrial-like 48.1 7e-06
ath:AT5G47840 AMK2; AMK2 (Adenosine monophosphate kinase); ATP... 47.8 8e-06
cel:ZK673.2 hypothetical protein; K00939 adenylate kinase [EC:... 47.4 1e-05
ath:AT5G35170 adenylate kinase family protein (EC:2.7.4.3); K0... 47.0 1e-05
mmu:68870 Ak8, 1190002A17Rik, MGC118311, MGC130365; adenylate ... 40.8 0.001
dre:553671 MGC112030, fb81g03, si:ch211-89p1.4, wu:fb81g03; zg... 39.3 0.003
hsa:158067 AK8, C9orf98, DDX31, FLJ32704, FLJ36014, RP11-143F1... 38.5 0.005
xla:496205 ak8, c9orf98, ddx31; adenylate kinase 8 (EC:2.7.4.3) 36.6 0.021
tgo:TGME49_058640 adenylate kinase, putative (EC:2.7.4.3); K00... 32.3 0.36
tgo:TGME49_081970 adenylate kinase, putative (EC:2.7.4.3); K00... 31.6 0.74
ath:AT2G12190 cytochrome P450, putative 29.6 2.2
ath:AT1G64950 CYP89A5; electron carrier/ heme binding / iron i... 28.9 4.4
ath:AT1G64940 CYP89A6; electron carrier/ heme binding / iron i... 28.5 5.2
ath:AT4G25280 adenylate kinase family protein (EC:2.7.4.14); K... 28.1 6.6
> tgo:TGME49_024900 adenylate kinase, putative (EC:2.7.4.10);
K00939 adenylate kinase [EC:2.7.4.3]
Length=257
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 35/57 (61%), Positives = 42/57 (73%), Gaps = 0/57 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
LD LL + QKLDGV++FD P+ LV+R+ GRRIH SGRVYH + PPKV G DDV
Sbjct 132 LDNLLKQKNQKLDGVLYFDVPDNILVERVSGRRIHLPSGRVYHVTYHPPKVAGLDDV 188
> pfa:PF10_0086 adenylate kinase; K00939 adenylate kinase [EC:2.7.4.3]
Length=242
Score = 77.8 bits (190), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 34/60 (56%), Positives = 44/60 (73%), Gaps = 0/60 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDVRHD 86
L+KLL + KLDGV +F+ P+E LV RI GR IH SGR+YH +F PPKVP +DDV ++
Sbjct 124 LNKLLQKNQTKLDGVFYFNVPDEVLVNRISGRLIHKPSGRIYHKIFNPPKVPFRDDVTNE 183
> xla:379924 ak2, MGC53010; adenylate kinase 2 (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=241
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/63 (52%), Positives = 42/63 (66%), Gaps = 0/63 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDVRHD 86
LD+LL +++KLD V+ F + LV+RICGR IH +SGR YH F PPK P KDDV +
Sbjct 113 LDELLEKRQEKLDSVIEFKVDDSLLVRRICGRLIHASSGRSYHEEFNPPKEPMKDDVTGE 172
Query 87 CSI 89
I
Sbjct 173 ALI 175
> tpv:TP01_0633 adenylate kinase; K00939 adenylate kinase [EC:2.7.4.3]
Length=245
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/71 (46%), Positives = 44/71 (61%), Gaps = 0/71 (0%)
Query 13 LFRWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVF 72
L + + S L+KLL S +KL+GV F+ +E + +RI GR +HP S RVYH VF
Sbjct 109 LLDGYPRSISQAKDLNKLLKSVGKKLNGVFSFNVSDEVIEKRIVGRLVHPGSNRVYHKVF 168
Query 73 RPPKVPGKDDV 83
PPK GKDD+
Sbjct 169 NPPKEEGKDDL 179
> mmu:11637 Ak2, Ak-2, D4Ertd220e; adenylate kinase 2 (EC:2.7.4.3);
K00939 adenylate kinase [EC:2.7.4.3]
Length=239
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 30/57 (52%), Positives = 38/57 (66%), Gaps = 0/57 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
LD L+ +K+KLD V+ F + L++RI GR IHP SGR YH F PPK P KDD+
Sbjct 111 LDDLMEKRKEKLDSVIEFSIQDSLLIRRITGRLIHPKSGRSYHEEFNPPKEPMKDDI 167
> eco:b0474 adk, dnaW, ECK0468, JW0463, plsA; adenylate kinase
(EC:2.7.4.3); K00939 adenylate kinase [EC:2.7.4.3]
Length=214
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/47 (61%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+D V+ FD P+E +V RI GRR+H SGRVYH F PPKV GKDDV
Sbjct 102 NVDYVLEFDVPDELIVDRIVGRRVHAPSGRVYHVKFNPPKVEGKDDV 148
> ath:AT5G50370 adenylate kinase, putative (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=248
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 40/58 (68%), Gaps = 0/58 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+LD++L + ++D V+ F + L +RI GR IHP+SGR YHT F PPKVPG DD+
Sbjct 129 KLDEMLNRRGAQIDKVLNFAIDDSVLEERITGRWIHPSSGRSYHTKFAPPKVPGVDDL 186
> cel:C29E4.8 let-754; LEThal family member (let-754); K00939
adenylate kinase [EC:2.7.4.3]
Length=251
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+LD++L +K LD VV F+ ++ LV+RI GR H ASGR YH F+PPKVP KDD+
Sbjct 121 KLDEILERRKTPLDTVVEFNIADDLLVRRITGRLFHIASGRSYHLEFKPPKVPMKDDL 178
> dre:321793 ak2, wu:fb34f05, wu:fj80e03; adenylate kinase 2 (EC:2.7.4.3);
K00939 adenylate kinase [EC:2.7.4.3]
Length=241
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 30/57 (52%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
LD L+ + +KLD V+ F + LV+RICGR IH SGR YH F PPK KDDV
Sbjct 113 LDDLMEKRSEKLDSVIEFSVDDSLLVRRICGRLIHQPSGRSYHEEFHPPKEHMKDDV 169
> sce:YDR226W ADK1, AKY1, AKY2; Adenylate kinase, required for
purine metabolism; localized to the cytoplasm and the mitochondria;
lacks cleavable signal sequence (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=222
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 37/62 (59%), Gaps = 0/62 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDVRH 85
+LD++L Q L+ + +E LV RI GR IHPASGR YH +F PPK KDDV
Sbjct 102 KLDQMLKEQGTPLEKAIELKVDDELLVARITGRLIHPASGRSYHKIFNPPKEDMKDDVTG 161
Query 86 DC 87
+
Sbjct 162 EA 163
> cpv:cgd5_3360 adenylate kinase ; K00939 adenylate kinase [EC:2.7.4.3]
Length=221
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
L K+L+ L V++F+ + +++RI GR HPASGR+YH + PPK PG DDV
Sbjct 103 LAKILSEIGDSLTSVIYFEIDDSEIIERISGRCTHPASGRIYHVKYNPPKQPGIDDV 159
> sce:YER170W ADK2, AKY3, PAK3; Adk2p (EC:2.7.4.3); K00939 adenylate
kinase [EC:2.7.4.3]
Length=225
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 42/74 (56%), Gaps = 0/74 (0%)
Query 10 ALRLFRWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYH 69
A+ L F + LD+LL L+ VV D PE T+++RI R +H SGRVY+
Sbjct 97 AMWLLDGFPRTTAQASALDELLKQHDASLNLVVELDVPESTILERIENRYVHVPSGRVYN 156
Query 70 TVFRPPKVPGKDDV 83
+ PPKVPG DD+
Sbjct 157 LQYNPPKVPGLDDI 170
> ath:AT5G63400 ADK1; ADK1 (ADENYLATE KINASE 1); ATP binding /
adenylate kinase/ nucleobase, nucleoside, nucleotide kinase/
nucleotide kinase/ phosphotransferase, phosphate group as
acceptor; K00939 adenylate kinase [EC:2.7.4.3]
Length=190
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+LD++L + ++D V+ F + L +RI GR IHP+SGR YHT F PPK PG DD+
Sbjct 128 KLDEMLKRRGTEIDKVLNFAIDDAILEERITGRWIHPSSGRSYHTKFAPPKTPGVDDM 185
> bbo:BBOV_IV002930 21.m02778; adenylate kinase 2 (EC:2.7.4.3);
K00939 adenylate kinase [EC:2.7.4.3]
Length=259
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDVRH 85
+L LL++ ++L+ V F+ P++ + +RI GR +H SGRVYH +PPKVP +DD+ +
Sbjct 136 RLKTLLSNLGKRLNAVFLFECPDDEIQRRITGRLVHEPSGRVYHMTSKPPKVPMRDDITN 195
Query 86 D 86
+
Sbjct 196 E 196
> xla:734496 ak3, MGC114646, ak6; adenylate kinase 3 (EC:2.7.4.10)
Length=221
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 24/46 (52%), Positives = 33/46 (71%), Gaps = 0/46 (0%)
Query 38 LDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++ V+ + P +T+ R+ R IHPASGRVY+T F PPKVPG DD+
Sbjct 107 INSVIDLNVPFQTIKDRLTARWIHPASGRVYNTEFNPPKVPGVDDL 152
> hsa:204 AK2, ADK2, AK_2; adenylate kinase 2 (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=224
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 33/57 (57%), Gaps = 8/57 (14%)
Query 27 LDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
LD L+ +K+KLD V+ F P+ L IHP SGR YH F PPK P KDD+
Sbjct 111 LDDLMEKRKEKLDSVIEFSIPDSLL--------IHPKSGRSYHEEFNPPKEPMKDDI 159
> xla:414511 hypothetical protein MGC82246; K00944 nucleoside-triphosphate--adenylate
kinase [EC:2.7.4.10]
Length=221
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 32/46 (69%), Gaps = 0/46 (0%)
Query 38 LDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++ V+ + P +T+ R+ R IHPASGRVY+T F PPKVP DD+
Sbjct 107 INSVIDLNVPFQTIKGRLTARWIHPASGRVYNTEFNPPKVPEVDDL 152
> mmu:56248 Ak3, AA407498, AI506714, AK-3, Ak3l, Ak3l1, Akl3l;
adenylate kinase 3 (EC:2.7.4.10); K00944 nucleoside-triphosphate--adenylate
kinase [EC:2.7.4.10]
Length=227
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++D V+ + P E + QR+ R IHPASGRVY+ F PPK G DD+
Sbjct 107 QIDTVINLNVPFEVIKQRLTARWIHPASGRVYNIEFNPPKTVGIDDL 153
> hsa:50808 AK3, AK3L1, AK6, AKL3L, AKL3L1, FIX; adenylate kinase
3 (EC:2.7.4.10); K00944 nucleoside-triphosphate--adenylate
kinase [EC:2.7.4.10]
Length=187
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++D V+ + P E + QR+ R IHPASGRVY+ F PPK G DD+
Sbjct 67 QIDTVINLNVPFEVIKQRLTARWIHPASGRVYNIEFNPPKTVGIDDL 113
> dre:406590 ak4, ak3l1, wu:fc37g02, zgc:85790; adenylate kinase
4 (EC:2.7.4.3); K00939 adenylate kinase [EC:2.7.4.3]
Length=226
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 41/82 (50%), Gaps = 1/82 (1%)
Query 8 FQALRLFRWFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRV 67
+ + + W + F + L S LD + + P ETL +R+ R IHP SGRV
Sbjct 76 LEEMTKYSWLLDGFPRTLAQAEALNSSCD-LDVAINLNIPLETLKERLRHRWIHPPSGRV 134
Query 68 YHTVFRPPKVPGKDDVRHDCSI 89
Y+ F PP++ G DD+ + I
Sbjct 135 YNMCFNPPRIQGLDDITGEALI 156
> mmu:11639 Ak4, Ak-3, Ak-4, Ak3, Ak3l1, D4Ertd274e; adenylate
kinase 4 (EC:2.7.4.3); K00939 adenylate kinase [EC:2.7.4.3]
Length=223
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 34/56 (60%), Gaps = 6/56 (10%)
Query 34 QKQKLDG------VVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
Q + LDG V+ + P ETL R+ R IHP+SGRVY+ F PP+V G DD+
Sbjct 96 QAEALDGICDVDLVISLNIPFETLKDRLSRRWIHPSSGRVYNLDFNPPQVQGIDDI 151
> dre:406404 ak3, ak3l1, id:ibd3034, sb:cb845, wu:fa02c11, wu:fc20h08,
zgc:64135; adenylate kinase 3 (EC:2.7.4.10); K00944
nucleoside-triphosphate--adenylate kinase [EC:2.7.4.10]
Length=214
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 38 LDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+D V+ D P +T+ +R+ R +H SGRVY+ F PPK PG DDV
Sbjct 96 VDSVINLDVPFQTIRERLTSRWVHLPSGRVYNIDFNPPKKPGLDDV 141
> hsa:205 AK4, AK3, AK3L1, AK3L2, MGC166959; adenylate kinase
4 (EC:2.7.4.3); K00939 adenylate kinase [EC:2.7.4.3]
Length=223
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++D V+ + P ETL R+ R IHP SGRVY+ F PP V G DDV
Sbjct 105 EVDLVISLNIPFETLKDRLSRRWIHPPSGRVYNLDFNPPHVHGIDDV 151
> hsa:100507855 adenylate kinase isoenzyme 4, mitochondrial-like
Length=223
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
++D V+ + P ETL R+ R IHP SGRVY+ F PP V G DDV
Sbjct 105 EVDLVISLNIPFETLKDRLSRRWIHPPSGRVYNLDFNPPHVHGIDDV 151
> ath:AT5G47840 AMK2; AMK2 (Adenosine monophosphate kinase); ATP
binding / adenylate kinase/ nucleobase, nucleoside, nucleotide
kinase/ nucleotide kinase/ phosphotransferase, phosphate
group as acceptor; K00939 adenylate kinase [EC:2.7.4.3]
Length=283
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 30/52 (57%), Gaps = 7/52 (13%)
Query 32 ASQKQKLDGVVF-------FDTPEETLVQRICGRRIHPASGRVYHTVFRPPK 76
ASQ L G F + PEE L++R+ GRR+ P +G++YH + PP+
Sbjct 155 ASQATALKGFGFQPDLFIVLEVPEEILIERVVGRRLDPVTGKIYHLKYSPPE 206
> cel:ZK673.2 hypothetical protein; K00939 adenylate kinase [EC:2.7.4.3]
Length=222
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 31 LASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDV 83
+ ++ L+ +V P + L+ R+ + +HPASGR Y+ PPK GKDD+
Sbjct 97 MVEEQAPLNLIVELKVPRKVLIDRLSKQLVHPASGRAYNLEVNPPKEEGKDDI 149
> ath:AT5G35170 adenylate kinase family protein (EC:2.7.4.3);
K00939 adenylate kinase [EC:2.7.4.3]
Length=580
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 16 WFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPP 75
W + F F + L K D + D P+E L+ R GRR+ P +G++YH PP
Sbjct 161 WLLDGFPRSFAQAQSLDKLNVKPDIFILLDVPDEILIDRCVGRRLDPVTGKIYHIKNYPP 220
Query 76 K 76
+
Sbjct 221 E 221
> mmu:68870 Ak8, 1190002A17Rik, MGC118311, MGC130365; adenylate
kinase 8 (EC:2.7.4.3)
Length=479
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 41 VVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVP 78
V+ + P+ L++R G+RI P +G +YHT F P P
Sbjct 161 VIVLNAPDTVLIERNVGKRIDPVTGEIYHTTFDWPPEP 198
> dre:553671 MGC112030, fb81g03, si:ch211-89p1.4, wu:fb81g03;
zgc:112030 (EC:2.7.4.3); K00939 adenylate kinase [EC:2.7.4.3]
Length=486
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 39 DGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPK 76
D VV + P+ L++R G+RI P +G VYH F P+
Sbjct 158 DHVVMLEAPDVVLIERSSGKRIDPVTGDVYHVTFMWPE 195
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 16/63 (25%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 16 WFIFAFSLHFQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPP 75
W + F + +LL + V F + ++ ++R+ R + P +G YH+V++PP
Sbjct 350 WVLHGFPRDVEEAELLHKSNFTPNRVFFLEITDDIAIERLALRAVDPVTGEWYHSVYKPP 409
Query 76 KVP 78
P
Sbjct 410 PGP 412
> hsa:158067 AK8, C9orf98, DDX31, FLJ32704, FLJ36014, RP11-143F18.1;
adenylate kinase 8 (EC:2.7.4.3)
Length=479
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 41 VVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPP 75
V F + P +++++R+ RRI P +G YH +++PP
Sbjct 375 VFFLNVPFDSIMERLTLRRIDPVTGERYHLMYKPP 409
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query 41 VVFFDTPEETLVQRICGRRIHPASGRVYHTVFR-PPK 76
V+ P+ L++R G+RI P +G +YHT F PP+
Sbjct 161 VIVLSAPDTVLIERNLGKRIDPQTGEIYHTTFDWPPE 197
> xla:496205 ak8, c9orf98, ddx31; adenylate kinase 8 (EC:2.7.4.3)
Length=485
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 41 VVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPG 79
+V D P+ L++R G+RI G VYHT F P P
Sbjct 161 LVVLDAPDIVLIERNMGKRIDITDGEVYHTTFDWPSDPA 199
> tgo:TGME49_058640 adenylate kinase, putative (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=375
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 5/49 (10%)
Query 37 KLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFRPPKVPGKDDVRH 85
K D +V D + L QR+ GR + P + +VY PP +++RH
Sbjct 204 KPDAIVCLDVDDAVLFQRLGGRLVDPETAQVYGAANPPP-----EEIRH 247
> tgo:TGME49_081970 adenylate kinase, putative (EC:2.7.4.3); K00939
adenylate kinase [EC:2.7.4.3]
Length=374
Score = 31.6 bits (70), Expect = 0.74, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 26 QLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVY 68
Q+D LL K D P TL QR+ RR+ P +G+V+
Sbjct 227 QVD-LLREMKLWPDVCFLLKVPNATLFQRMAARRVDPVTGQVF 268
> ath:AT2G12190 cytochrome P450, putative
Length=512
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR 73
F D+ LA Q L+G VF D P + +I H S +Y +R
Sbjct 81 FVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSSLYGATWR 129
> ath:AT1G64950 CYP89A5; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=510
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR 73
F D+ LA Q L+G VF D P + +I H S +Y +R
Sbjct 81 FVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSCLYGATWR 129
> ath:AT1G64940 CYP89A6; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=511
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 0/49 (0%)
Query 25 FQLDKLLASQKQKLDGVVFFDTPEETLVQRICGRRIHPASGRVYHTVFR 73
F D+ LA Q L+G VF D P + +I H S +Y +R
Sbjct 82 FVTDRSLAHQALVLNGAVFADRPPAESISKIISSNQHNISSCLYGATWR 130
> ath:AT4G25280 adenylate kinase family protein (EC:2.7.4.14);
K13800 UMP-CMP kinase [EC:2.7.4.- 2.7.4.14]
Length=249
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 39 DGVVFFDTPEETLVQRICGRR 59
D V+FFD PEE +V+R+ R
Sbjct 147 DVVLFFDCPEEEMVKRVLNRN 167
Lambda K H
0.332 0.147 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2074765676
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40