bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2774_orf1
Length=133
Score E
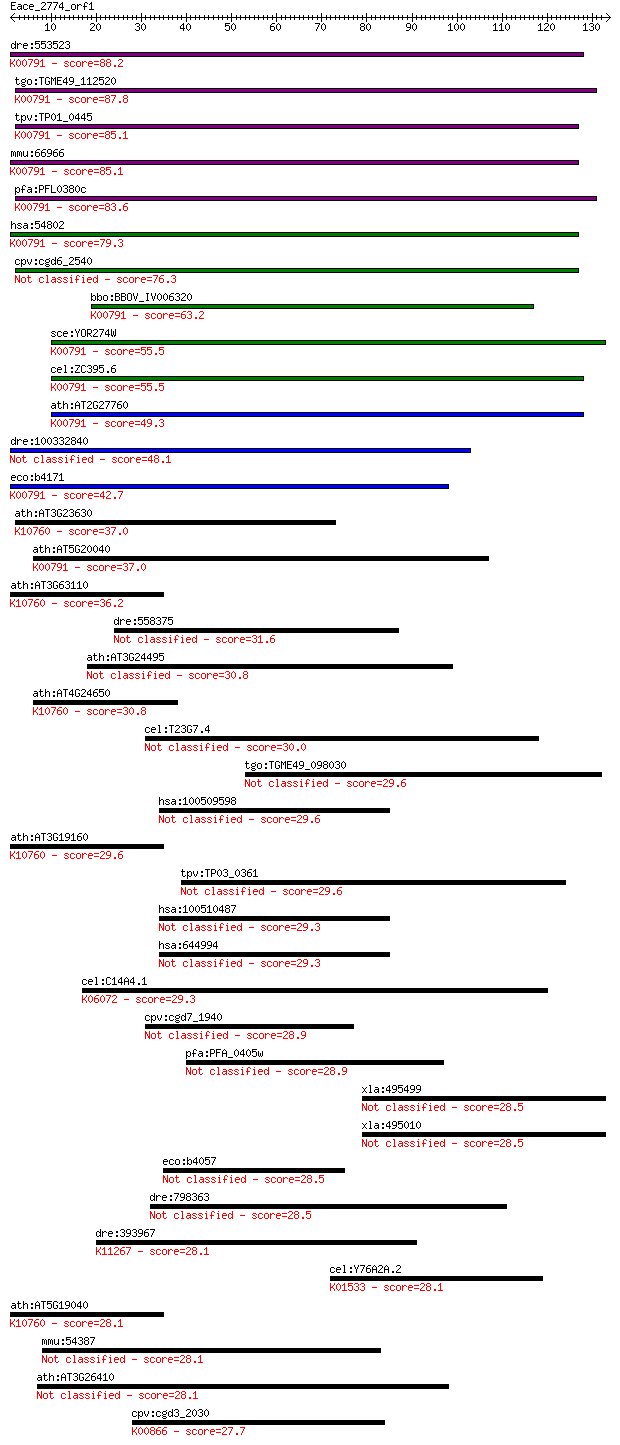
Sequences producing significant alignments: (Bits) Value
dre:553523 trit1, si:ch211-194e15.1; tRNA isopentenyltransfera... 88.2 6e-18
tgo:TGME49_112520 tRNA delta(2)-isopentenylpyrophosphate trans... 87.8 7e-18
tpv:TP01_0445 tRNA delta(2)-isopentenylpyrophosphate transfera... 85.1 5e-17
mmu:66966 Trit1, 2310075G14Rik, AI314189, AI314635, AV099619, ... 85.1 6e-17
pfa:PFL0380c tRNA delta(2)-isopentenylpyrophosphate transferas... 83.6 2e-16
hsa:54802 TRIT1, FLJ20061, IPT, MGC149242, MGC149243, MOD5; tR... 79.3 3e-15
cpv:cgd6_2540 tRNA delta(2)-isopentenylpyrophosphate transferase 76.3 2e-14
bbo:BBOV_IV006320 23.m06027; hypothetical protein; K00791 tRNA... 63.2 2e-10
sce:YOR274W MOD5; Delta 2-isopentenyl pyrophosphate:tRNA isope... 55.5 4e-08
cel:ZC395.6 gro-1; abnormal GROwth rate family member (gro-1);... 55.5 5e-08
ath:AT2G27760 ATIPT2; ATIPT2 (TRNA ISOPENTENYLTRANSFERASE 2); ... 49.3 3e-06
dre:100332840 tRNA delta(2)-isopentenylpyrophosphate transfera... 48.1 7e-06
eco:b4171 miaA, ECK4167, JW4129, trpX; delta(2)-isopentenylpyr... 42.7 3e-04
ath:AT3G23630 ATIPT7; ATIPT7; ATP binding / tRNA isopentenyltr... 37.0 0.015
ath:AT5G20040 ATIPT9; ATIPT9; ATP binding / tRNA isopentenyltr... 37.0 0.016
ath:AT3G63110 ATIPT3; ATIPT3 (ARABIDOPSIS THALIANA ISOPENTENYL... 36.2 0.031
dre:558375 finTRIM family, member 72-like 31.6 0.68
ath:AT3G24495 MSH7; MSH7 (MUTS HOMOLOG 7); ATP binding / damag... 30.8 1.1
ath:AT4G24650 ATIPT4; ATIPT4; adenylate dimethylallyltransfera... 30.8 1.3
cel:T23G7.4 sec-5; yeast SEC homolog family member (sec-5) 30.0 1.8
tgo:TGME49_098030 hypothetical protein 29.6 2.6
hsa:100509598 hypothetical protein LOC100509598 29.6 2.8
ath:AT3G19160 ATIPT8; ATIPT8 (ATP/ADP ISOPENTENYLTRANSFERASES)... 29.6 2.8
tpv:TP03_0361 hypothetical protein 29.6 2.9
hsa:100510487 hypothetical protein LOC100510487 29.3 3.0
hsa:644994 C5orf50; chromosome 5 open reading frame 50 29.3
cel:C14A4.1 tag-242; Temporarily Assigned Gene name family mem... 29.3 3.4
cpv:cgd7_1940 coiled coil protein 28.9 3.9
pfa:PFA_0405w conserved Plasmodium protein, unknown function 28.9 4.1
xla:495499 tnpo2; transportin 2 28.5
xla:495010 hypothetical LOC495010 28.5 6.1
eco:b4057 yjbR, ECK4049, JW4018; conserved protein 28.5 6.2
dre:798363 hypothetical LOC798363 28.5 6.3
dre:393967 pds5a, Aprin, MGC153878, MGC66331, zgc:153878, zgc:... 28.1 6.8
cel:Y76A2A.2 cua-1; CU (copper) ATPase family member (cua-1); ... 28.1 7.6
ath:AT5G19040 IPT5; IPT5; ATP binding / tRNA isopentenyltransf... 28.1 7.8
mmu:54387 Mcm3ap, GANP, mKIAA0572; minichromosome maintenance ... 28.1 8.4
ath:AT3G26410 methyltransferase/ nucleic acid binding 28.1 8.6
cpv:cgd3_2030 choline kinase ; K00866 choline kinase [EC:2.7.1... 27.7 8.9
> dre:553523 trit1, si:ch211-194e15.1; tRNA isopentenyltransferase
1 (EC:2.5.1.75); K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=447
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 51/140 (36%), Positives = 75/140 (53%), Gaps = 16/140 (11%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIA----T 56
D+ A I+ + + KLPV+VGGT YI+ +LWK +D Q ++ E +
Sbjct 88 DFRNKALSLIEDMHHRKKLPVIVGGTNYYIESILWKVLIDTG---QGSDTEPEKGGGADS 144
Query 57 REALEAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSE- 115
R LE EL LK++D A LHPHD R+I RS++V + GVPHS +++ + +
Sbjct 145 RAGLEKLGGPELHRRLKEVDPDMAAILHPHDSRKIARSLQVYMDTGVPHSRLLEEQRGQD 204
Query 116 -------KLRY-DACIFWLD 127
LR+ D CIFWL+
Sbjct 205 GGDCLGGPLRFQDPCIFWLN 224
> tgo:TGME49_112520 tRNA delta(2)-isopentenylpyrophosphate transferase,
putative (EC:2.5.1.8); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=741
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 52/135 (38%), Positives = 74/135 (54%), Gaps = 23/135 (17%)
Query 2 YLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNK--QNTNITKEIATREA 59
+L+ A QTI+ L +G LP+VVGGTQLY+Q LLW+S +D +++ ++ K +
Sbjct 254 FLQLATQTIEALHARGVLPIVVGGTQLYLQHLLWRSLLDRYLHEDLESEMQDKSSGSCSP 313
Query 60 LEAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENS----E 115
L+ + E L LK++D +R TQ GVPHSE+M+ E E
Sbjct 314 LDGFSTETLYAELKKLDPER-----------------TTQSTGVPHSELMRREREASIRE 356
Query 116 KLRYDACIFWLDLRD 130
LRY +CI WLD RD
Sbjct 357 GLRYPSCILWLDCRD 371
> tpv:TP01_0445 tRNA delta(2)-isopentenylpyrophosphate transferase;
K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=374
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 49/133 (36%), Positives = 77/133 (57%), Gaps = 9/133 (6%)
Query 2 YLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREALE 61
++K I L + KLPV+VGGT LYI+ LLW S +D + N I ++ E+LE
Sbjct 113 FVKDCSHLINVLFSENKLPVIVGGTNLYIEALLWPSVMDSNHNSSLLFIL-FLSDSESLE 171
Query 62 AKTNE----ELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMK----GEN 113
+E L LL +ID +RA+QLH +DR+RI+RS+++ G+ H+ ++K ++
Sbjct 172 EYFDELDTRTLYNLLTEIDPERAKQLHQNDRKRILRSLDIYHSQGITHTNLIKLRKDQKH 231
Query 114 SEKLRYDACIFWL 126
E +RYD + L
Sbjct 232 KEGIRYDPLVLIL 244
> mmu:66966 Trit1, 2310075G14Rik, AI314189, AI314635, AV099619,
IPT, MOD5; tRNA isopentenyltransferase 1 (EC:2.5.1.75); K00791
tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=467
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 76/135 (56%), Gaps = 11/135 (8%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREAL 60
D+ A I+ + + K+P+VVGGT YI+ LLWK V I+ Q K + + L
Sbjct 97 DFRNKATALIEDIFARDKIPIVVGGTNYYIESLLWK--VLITTKPQEMGTGKVVDRKVEL 154
Query 61 EAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENS------ 114
E + EL + L Q+D + A +LHPHD+R++ RS++V +E G+ HSE + +++
Sbjct 155 EKEDGHELHKRLSQVDPEMAAKLHPHDKRKVARSLQVFEETGISHSEFLHRQHAEEGGGP 214
Query 115 --EKLRY-DACIFWL 126
LR+ + CI WL
Sbjct 215 LGGPLRFPNPCILWL 229
> pfa:PFL0380c tRNA delta(2)-isopentenylpyrophosphate transferase,
putative (EC:2.5.1.8); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=601
Score = 83.6 bits (205), Expect = 2e-16, Method: Composition-based stats.
Identities = 53/164 (32%), Positives = 83/164 (50%), Gaps = 35/164 (21%)
Query 2 YLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQ------NTNITK--- 52
Y+ I+ ++ K+P++ GGT LYI+ LLW+S +DI ++ N I K
Sbjct 308 YINYTIPLIKNMNRNNKIPIIAGGTLLYIESLLWESVIDIRKEEEKKEQTYNGQIIKKED 367
Query 53 ---------------------EIATREALEAKTNEELVELLKQIDSKRAEQLHPHDRRRI 91
++ + E KTNEEL E LK ID +RA QLH +DR+R+
Sbjct 368 EYLDEHLKLNNDIKCNGERNDDMVELDKYEHKTNEELYEELKNIDEERANQLHKNDRKRV 427
Query 92 IRSIEVTQEFGVPHSEVM-----KGENSEKLRYDACIFWLDLRD 130
RS+++ + HS+++ K N +K+R+ CIF+LD D
Sbjct 428 CRSLDIFYTYNKKHSDLIKIKNHKNNNIDKMRFFPCIFYLDYND 471
> hsa:54802 TRIT1, FLJ20061, IPT, MGC149242, MGC149243, MOD5;
tRNA isopentenyltransferase 1 (EC:2.5.1.75); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=467
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 46/135 (34%), Positives = 74/135 (54%), Gaps = 11/135 (8%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREAL 60
D+ A I+ + + K+P+VVGGT YI+ LLWK V ++ Q K I + L
Sbjct 97 DFRNRATALIEDIFARDKIPIVVGGTNYYIESLLWK--VLVNTKPQEMGTEKVIDRKVEL 154
Query 61 EAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKLRY- 119
E + L + L Q+D + A +LHPHD+R++ RS++V +E G+ HSE + +++E+
Sbjct 155 EKEDGLVLHKRLSQVDPEMAAKLHPHDKRKVARSLQVFEETGISHSEFLHRQHTEEGGGP 214
Query 120 --------DACIFWL 126
+ CI WL
Sbjct 215 LGGPLKFSNPCILWL 229
> cpv:cgd6_2540 tRNA delta(2)-isopentenylpyrophosphate transferase
Length=361
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/126 (35%), Positives = 75/126 (59%), Gaps = 6/126 (4%)
Query 2 YLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREALE 61
Y+ A I L + P++VGGT +Y++ L+WKS +D ++ +++ I E
Sbjct 72 YISIAVPIIDRLISENVTPIIVGGTHMYLKALIWKSIID---SETCSSVNPSIKDEEY-S 127
Query 62 AKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKLRY-D 120
TNEEL+E L +ID +RA L+ +DRR+IIR IE+ G+ +E +K + K+RY +
Sbjct 128 NFTNEELLEQLSKIDPERALSLNINDRRKIIRGIEIYNTNGLTFTE-LKKIHPYKVRYNN 186
Query 121 ACIFWL 126
+ +FW+
Sbjct 187 SVLFWV 192
> bbo:BBOV_IV006320 23.m06027; hypothetical protein; K00791 tRNA
dimethylallyltransferase [EC:2.5.1.75]
Length=185
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 62/107 (57%), Gaps = 11/107 (10%)
Query 19 LPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREA-------LEAKTNEELVEL 71
LP++VGGT LYI+ LLW S +D+ + QN+ T A L K+ +L +
Sbjct 75 LPIIVGGTNLYIEGLLWPSILDMRL--QNSETTDAYHGMTAHNNRITPLTGKSVVQLYDE 132
Query 72 LKQIDSKR--AEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEK 116
L ++D +R A LH +DR+RI RS++V + G+ HS+++ SEK
Sbjct 133 LMKLDPQRVMAAMLHCNDRKRITRSLDVIKTSGMKHSDLIAVRLSEK 179
> sce:YOR274W MOD5; Delta 2-isopentenyl pyrophosphate:tRNA isopentenyl
transferase, required for biosynthesis of the modified
base isopentenyladenosine in mitochondrial and cytoplasmic
tRNAs; gene is nuclear and encodes two isozymic forms (EC:2.5.1.8);
K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=428
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 59/123 (47%), Gaps = 6/123 (4%)
Query 10 IQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREALEAKTNEELV 69
I+ + +GK+P+VVGGT Y+Q L K S ++ T +I LE+ + +
Sbjct 96 IEDIHRRGKIPIVVGGTHYYLQTLFNKRVDTKSSERKLTRKQLDI-----LESTDPDVIY 150
Query 70 ELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKLRYDACIFWLDLR 129
L + D A + HP+D RR+ R +E+ + G SE E L++D WL +
Sbjct 151 NTLVKCDPDIATKYHPNDYRRVQRMLEIYYKTGKKPSETF-NEQKITLKFDTLFLWLYSK 209
Query 130 DRP 132
P
Sbjct 210 PEP 212
> cel:ZC395.6 gro-1; abnormal GROwth rate family member (gro-1);
K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=430
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 38/136 (27%), Positives = 78/136 (57%), Gaps = 24/136 (17%)
Query 10 IQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEI----------ATREA 59
I+ + + K+PV+VGGT Y + +L+++ N TN + ++ ++ +
Sbjct 102 IKKIRARSKIPVIVGGTTYYAESVLYEN------NLIETNTSDDVDSKSRTSSESSSEDT 155
Query 60 LEAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSE---- 115
E +N+EL + LK+ID K A LHP++R R+ R++++ +E G+ SE+++ + S+
Sbjct 156 EEGISNQELWDELKKIDEKSALLLHPNNRYRVQRALQIFRETGIRKSELVEKQKSDETVD 215
Query 116 ---KLRYD-ACIFWLD 127
+LR+D + + ++D
Sbjct 216 LGGRLRFDNSLVIFMD 231
> ath:AT2G27760 ATIPT2; ATIPT2 (TRNA ISOPENTENYLTRANSFERASE 2);
adenylate dimethylallyltransferase/ tRNA isopentenyltransferase
(EC:2.5.1.75); K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=466
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 67/137 (48%), Gaps = 25/137 (18%)
Query 10 IQTLSDQGKLPVVVGGTQLYIQLLLWKSAVD-------------ISVNKQNTNITKEIAT 56
I+ + + +PV+VGGT YIQ ++ K +D SV Q+ + + +
Sbjct 102 IEEIVSRNHIPVLVGGTHYYIQAVVSKFLLDDAAEDTEECCADVASVVDQDM-VVESVFG 160
Query 57 REALEAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEK 116
R+ L ELLK++D A ++HP++ R+I + + + GV S++ +G+ +E
Sbjct 161 RDDLSHG-----YELLKELDPVAANRIHPNNHRKINQYLSLHASRGVLPSKLYQGKTAEN 215
Query 117 L------RYDACIFWLD 127
R+D C+ +D
Sbjct 216 WGCINASRFDYCLICMD 232
> dre:100332840 tRNA delta(2)-isopentenylpyrophosphate transferase,
putative-like
Length=259
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 50/104 (48%), Gaps = 9/104 (8%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEI--ATRE 58
D+L+ + ++ L G+LPV VGGT LY + L + I EI R+
Sbjct 82 DWLRDVQASLPELRAGGRLPVFVGGTGLYFKALTGGLS-------DMPEIPAEIREGLRQ 134
Query 59 ALEAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFG 102
L E L L+ D AE+L+P D RI+R++EV + G
Sbjct 135 RLLDDGPEALHAELRACDPALAERLNPTDGHRIVRAVEVFRATG 178
> eco:b4171 miaA, ECK4167, JW4129, trpX; delta(2)-isopentenylpyrophosphate
tRNA-adenosine transferase (EC:2.5.1.75); K00791
tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=316
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 53/97 (54%), Gaps = 5/97 (5%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREAL 60
D+ + A + ++ G++P++VGGT LY + LL + S + + ++ A +
Sbjct 83 DFRRDALAEMADITAAGRIPLLVGGTMLYFKALLEGLSPLPSADPEVRARIEQQAAEQGW 142
Query 61 EAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEV 97
E+ + L+++D A ++HP+D +R+ R++EV
Sbjct 143 ESLHRQ-----LQEVDPVAAARIHPNDPQRLSRALEV 174
> ath:AT3G23630 ATIPT7; ATIPT7; ATP binding / tRNA isopentenyltransferase/
transferase, transferring alkyl or aryl (other
than methyl) groups; K10760 adenylate isopentenyltransferase
(cytokinin synthase)
Length=329
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 41/84 (48%), Gaps = 15/84 (17%)
Query 2 YLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSA-------------VDISVNKQNT 48
Y + A Q I LS KLP+V GG+ YI+ L+ S+ VD+S+ N+
Sbjct 109 YSRLASQAISKLSANNKLPIVAGGSNSYIEALVNHSSGFLLNNYDCCFIWVDVSLPVLNS 168
Query 49 NITKEIATREALEAKTNEELVELL 72
++K + +EA EE+ E+
Sbjct 169 FVSKRV--DRMMEAGLLEEVREVF 190
> ath:AT5G20040 ATIPT9; ATIPT9; ATP binding / tRNA isopentenyltransferase
(EC:2.5.1.27); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=459
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 26/103 (25%), Positives = 50/103 (48%), Gaps = 5/103 (4%)
Query 6 AKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATR-EALEAKT 64
+Q + + ++G++P+V GGT LY++ ++ V K + + E + +
Sbjct 128 GRQATKDILNRGRVPIVTGGTGLYLRWFMYGKP---DVPKPSPEVIAEAHDMLVGFQTEY 184
Query 65 N-EELVELLKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHS 106
N + VEL+ +A L +D R+ RS+E+ + G P S
Sbjct 185 NWDAAVELVVNAGDPKASSLPRNDWYRLRRSLEILKSTGSPPS 227
> ath:AT3G63110 ATIPT3; ATIPT3 (ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE
3); ATP binding / tRNA isopentenyltransferase/
transferase, transferring alkyl or aryl (other than methyl)
groups; K10760 adenylate isopentenyltransferase (cytokinin
synthase)
Length=336
Score = 36.2 bits (82), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLL 34
+Y A +I+++ ++GKLP++VGG+ Y++ L+
Sbjct 114 NYCHMANLSIESVLNRGKLPIIVGGSNSYVEALV 147
> dre:558375 finTRIM family, member 72-like
Length=537
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 24 GGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQL 83
G +L I + L+KS+ D +V K T+ I + E ++ E+ + ++ ++ RAEQL
Sbjct 205 GWQELSIAMKLFKSSADEAVEKMEKVFTELICSLEKKRSEIKEQ-IRAQEKTETDRAEQL 263
Query 84 HPH 86
H H
Sbjct 264 HQH 266
> ath:AT3G24495 MSH7; MSH7 (MUTS HOMOLOG 7); ATP binding / damaged
DNA binding / mismatched DNA binding
Length=1109
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 43/82 (52%), Gaps = 8/82 (9%)
Query 18 KLPVVVGGTQLYIQLLLWKSAVDISV-NKQNTNITKEIATREALEAKTNEELVELLKQID 76
KLP++VG + L + L +++A+D N QN ++T E A+T L+EL +
Sbjct 704 KLPILVGKSGLELFLSQFEAAIDSDFPNYQNQDVTDE-------NAETLTILIELFIERA 756
Query 77 SKRAEQLHPHDRRRIIRSIEVT 98
++ +E +H ++RS +
Sbjct 757 TQWSEVIHTISCLDVLRSFAIA 778
> ath:AT4G24650 ATIPT4; ATIPT4; adenylate dimethylallyltransferase;
K10760 adenylate isopentenyltransferase (cytokinin synthase)
Length=318
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 6 AKQTIQTLSDQGKLPVVVGGTQLYIQLLLWKS 37
A + I ++ + KLP++ GG+ YI LL KS
Sbjct 84 AAEAISEITQRKKLPILAGGSNSYIHALLAKS 115
> cel:T23G7.4 sec-5; yeast SEC homolog family member (sec-5)
Length=884
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 44/93 (47%), Gaps = 9/93 (9%)
Query 31 QLLLWKSAVDISVNKQNTNITKEIATREALEAKTNEEL------VELLKQIDSKRAEQLH 84
Q+LL + D S N + N + + E T E+L +EL KQ ++KR+E++H
Sbjct 148 QVLLSRDFPDCSGNLRMENFSPQWYLLENHADATIEDLRIAIKNMELSKQNEAKRSEEMH 207
Query 85 PHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKL 117
+ +I ++ H + KGEN++
Sbjct 208 KANLYSLINCVDTLANL---HQALEKGENADHF 237
> tgo:TGME49_098030 hypothetical protein
Length=670
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 37/85 (43%), Gaps = 21/85 (24%)
Query 53 EIATREALEAKTNEELVELLKQIDSKRAEQ---LHPHDRRRI---IRSIEVTQEFGVPHS 106
E+A +L A NE L++L K D+ AE+ HP +RI ++SI + G
Sbjct 219 EVAAAGSLGAAINERLIDLYKNSDAGAAERKKTYHPVRCQRIAVMLKSIYAVLDRG---- 274
Query 107 EVMKGENSEKLRYDACIFWLDLRDR 131
+ C+FW D + R
Sbjct 275 -----------SFVHCLFWFDRKHR 288
> hsa:100509598 hypothetical protein LOC100509598
Length=371
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 34 LWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQLH 84
+W S+ ++S + N + +A + LE +TNE E +K I + E+LH
Sbjct 190 IWGSSWEVSERIRECNYYQNLAVPQGLEYQTNEPSEEPIKTIRNWLKEKLH 240
> ath:AT3G19160 ATIPT8; ATIPT8 (ATP/ADP ISOPENTENYLTRANSFERASES);
adenylate dimethylallyltransferase (EC:2.5.1.27); K10760
adenylate isopentenyltransferase (cytokinin synthase)
Length=330
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 10/34 (29%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLL 34
++ A ++I ++ +G LP++ GG+ +I LL
Sbjct 117 EFRSLASRSISEITARGNLPIIAGGSNSFIHALL 150
> tpv:TP03_0361 hypothetical protein
Length=315
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 39/85 (45%), Gaps = 6/85 (7%)
Query 39 VDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQLHPHDRRRIIRSIEVT 98
+D +NK N E AT E + +LLKQ+++ + EQ+ H + ++ T
Sbjct 235 MDCLLNKYLANTQDENATGNPPEDNYYSQ--QLLKQMENNKTEQILTHKVNAKVYTLGFT 292
Query 99 QEFGVPHSEVMKGENSEKLRYDACI 123
+ G+ + E KL+++ C
Sbjct 293 EVNGIEYPE----NEPAKLKFNTCF 313
> hsa:100510487 hypothetical protein LOC100510487
Length=337
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 34 LWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQLH 84
+W S+ ++S + N + +A + LE +TNE E +K I + E+LH
Sbjct 156 IWGSSWEVSERIRECNYYQNLAVPQGLEYQTNEPSEEPIKTIRNWLKEKLH 206
> hsa:644994 C5orf50; chromosome 5 open reading frame 50
Length=337
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 34 LWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQLH 84
+W S+ ++S + N + +A + LE +TNE E +K I + E+LH
Sbjct 156 IWGSSWEVSERIRECNYYQNLAVPQGLEYQTNEPSEEPIKTIRNWLKEKLH 206
> cel:C14A4.1 tag-242; Temporarily Assigned Gene name family member
(tag-242); K06072 deoxyhypusine monooxygenase [EC:1.14.99.29]
Length=298
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 47/104 (45%), Gaps = 28/104 (26%)
Query 17 GKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQID 76
G+L V +L +LLL + +N + E A EAL A NEE E+LKQ
Sbjct 219 GQLQSPVATQELKDRLLL---------STENCMVRHECA--EALGAIANEECTEILKQ-- 265
Query 77 SKRAEQLHPHDRRRIIR-SIEVTQEFGVPHSEVMKGENSEKLRY 119
+ +D R++R S EV + + ENS+ L+Y
Sbjct 266 -------YVNDEERVVRESCEVALDMA-------EYENSDDLQY 295
> cpv:cgd7_1940 coiled coil protein
Length=564
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 5/51 (9%)
Query 31 QLLLWKSAVDISVNKQNTNITKEI-----ATREALEAKTNEELVELLKQID 76
QL K A+++S NK N I +E E + +++E+LVELLK+ID
Sbjct 424 QLEETKKALEVSQNKYNNLIKQESINAKWPNYELTKGRSHEDLVELLKKID 474
> pfa:PFA_0405w conserved Plasmodium protein, unknown function
Length=656
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Query 40 DISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQLH-PHDRRRIIRSIE 96
DI+VN N N T+ ++ + + K +E+ V L KQI ++ Q+ PH +II I+
Sbjct 114 DINVNDTNINSTQNTSSNKVMSCK-DEKYVILKKQITNEFIHQVFLPHVESQIIYHIK 170
> xla:495499 tnpo2; transportin 2
Length=890
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 25/54 (46%), Gaps = 12/54 (22%)
Query 79 RAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKLRYDACIFWLDLRDRP 132
R ++L PH I ++ TQ+ N E + +AC FWL L D+P
Sbjct 244 RIDRLLPHMHSIIQYMLQRTQD------------NDENVSLEACEFWLTLADQP 285
> xla:495010 hypothetical LOC495010
Length=889
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 25/54 (46%), Gaps = 12/54 (22%)
Query 79 RAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKLRYDACIFWLDLRDRP 132
R ++L PH I ++ TQ+ N E + +AC FWL L D+P
Sbjct 244 RIDRLLPHMHSIIQYMLQRTQD------------NDENVALEACEFWLTLADQP 285
> eco:b4057 yjbR, ECK4049, JW4018; conserved protein
Length=118
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query 35 WKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQ 74
WK A I V + KE+ R A+ KT+ EL ELL+Q
Sbjct 24 WK-ATQIKVEDVLFAMVKEVENRPAVSLKTSPELAELLRQ 62
> dre:798363 hypothetical LOC798363
Length=594
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 16/79 (20%), Positives = 39/79 (49%), Gaps = 2/79 (2%)
Query 32 LLLWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQLHPHDRRRI 91
L +W ++ N+ ++ I A AK E+V ++ +D R +L+P + +
Sbjct 119 LTVWPENFEVPWNRMSSGIKTATALGRRPTAKERREMVRII--VDEMRQTELNPSKSQCL 176
Query 92 IRSIEVTQEFGVPHSEVMK 110
I + ++ +++ ++V+K
Sbjct 177 IVAKKIVKQYPQSFADVLK 195
> dre:393967 pds5a, Aprin, MGC153878, MGC66331, zgc:153878, zgc:66331;
PDS5, regulator of cohesion maintenance, homolog A
(S. cerevisiae); K11267 sister chromatid cohesion protein PDS5
Length=1320
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 37/75 (49%), Gaps = 6/75 (8%)
Query 20 PVVVGGTQLYIQLLLWKSAVDISVNKQ----NTNITKEIATREALEAKTNEELVELLKQI 75
P+ V + YI+ ++ + S N Q TN ++++++ + A+ NEE + K +
Sbjct 1146 PLTVTARRPYIKTFTSETGSNASTNSQPSSPATNKSRDVSSE--VGARENEENPVITKAV 1203
Query 76 DSKRAEQLHPHDRRR 90
K+ E P R+R
Sbjct 1204 SVKKEEAAQPSGRKR 1218
> cel:Y76A2A.2 cua-1; CU (copper) ATPase family member (cua-1);
K01533 Cu2+-exporting ATPase [EC:3.6.3.4]
Length=1238
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 72 LKQIDSKRAEQLHPHDRRRIIRSIEVTQEFGVPHSEVMKGENSEKLR 118
L Q+ SK Q +P +I + + + G+P SEV+K SE+ R
Sbjct 902 LLQVSSKEVSQPNPDTANIVIGTERMMERHGIPVSEVVKMTLSEEQR 948
> ath:AT5G19040 IPT5; IPT5; ATP binding / tRNA isopentenyltransferase/
transferase, transferring alkyl or aryl (other than
methyl) groups; K10760 adenylate isopentenyltransferase (cytokinin
synthase)
Length=330
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 8/34 (23%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 1 DYLKAAKQTIQTLSDQGKLPVVVGGTQLYIQLLL 34
D+ + A + ++++ + ++P++ GG+ YI+ L+
Sbjct 107 DFQREAIRAVESIVQRDRVPIIAGGSNSYIEALV 140
> mmu:54387 Mcm3ap, GANP, mKIAA0572; minichromosome maintenance
deficient 3 (S. cerevisiae) associated protein
Length=1971
Score = 28.1 bits (61), Expect = 8.4, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 38/76 (50%), Gaps = 1/76 (1%)
Query 8 QTIQTLSDQGKLPVVVGGTQL-YIQLLLWKSAVDISVNKQNTNITKEIATREALEAKTNE 66
Q Q ++ +LP+ + T + + + ++ V S + QN+ K++ EA + E
Sbjct 1880 QDSQAFTESTRLPLYLPQTLVSFPDSIKTQTMVKTSTSPQNSGTGKQLRFSEASGSSLTE 1939
Query 67 ELVELLKQIDSKRAEQ 82
+L L + I S RAE+
Sbjct 1940 KLKLLERLIQSSRAEE 1955
> ath:AT3G26410 methyltransferase/ nucleic acid binding
Length=477
Score = 28.1 bits (61), Expect = 8.6, Method: Composition-based stats.
Identities = 22/91 (24%), Positives = 36/91 (39%), Gaps = 0/91 (0%)
Query 7 KQTIQTLSDQGKLPVVVGGTQLYIQLLLWKSAVDISVNKQNTNITKEIATREALEAKTNE 66
K +IQ+ D KLP + + I + + A+ K I + K +
Sbjct 88 KDSIQSYPDSRKLPFLTSDSTFRISVETFGKALTFDEQKDRIQSFTYIPFEGRVNLKNPD 147
Query 67 ELVELLKQIDSKRAEQLHPHDRRRIIRSIEV 97
L++ I+S+ L P +RRI EV
Sbjct 148 HNFFLMEMIESEENNGLQPILQRRIFFGREV 178
> cpv:cgd3_2030 choline kinase ; K00866 choline kinase [EC:2.7.1.32]
Length=405
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 28 LYIQLLLWKSAVDISVNKQNTNITKEIATREALEAKTNEELVELLKQIDSKRAEQL 83
L+ ++ LW+ I V+K N I KE+ ++ E EEL+ ++ +RA +L
Sbjct 179 LFKRIYLWREEAKIQVSKNNFQIDKELYSKILEEIDQLEELIMGGEKFSMERALEL 234
Lambda K H
0.315 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40