bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2778_orf1
Length=77
Score E
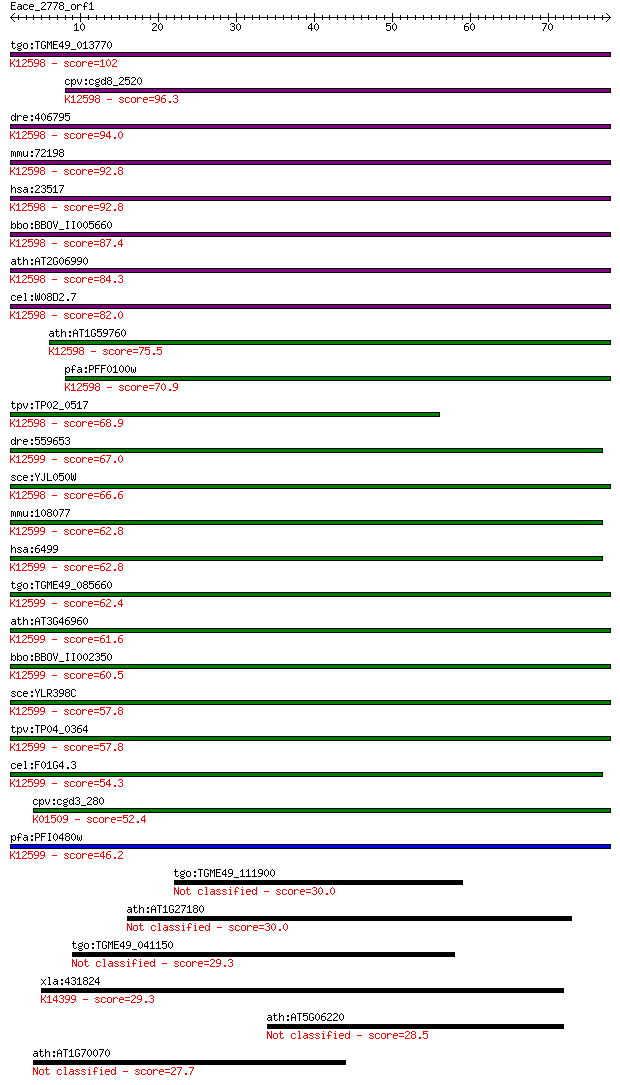
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_013770 RNA helicase, putative ; K12598 ATP-dependen... 102 2e-22
cpv:cgd8_2520 Mtr4p like SKI family SFII helicase ; K12598 ATP... 96.3 2e-20
dre:406795 skiv2l2, wu:fd11a05, zgc:63838; superkiller viralic... 94.0 1e-19
mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viral... 92.8 2e-19
hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118; s... 92.8 2e-19
bbo:BBOV_II005660 18.m06470; DSHCT (NUC185) domain containing ... 87.4 1e-17
ath:AT2G06990 HEN2; HEN2 (hua enhancer 2); ATP-dependent helic... 84.3 8e-17
cel:W08D2.7 mtr-4; yeast MTR (mRNA TRansport) homolog family m... 82.0 4e-16
ath:AT1G59760 ATP-dependent RNA helicase, putative; K12598 ATP... 75.5 4e-14
pfa:PFF0100w ATP-dependent RNA Helicase, putative (EC:3.6.1.-)... 70.9 9e-13
tpv:TP02_0517 hypothetical protein; K12598 ATP-dependent RNA h... 68.9 4e-12
dre:559653 skiv2l, fb70b07, wu:fb70b07; superkiller viralicidi... 67.0 1e-11
sce:YJL050W MTR4, DOB1; ATP-dependent 3'-5' RNA helicase, invo... 66.6 2e-11
mmu:108077 Skiv2l, 4930534J06Rik, AW214248, Ddx13, SKI, Ski2w;... 62.8 3e-10
hsa:6499 SKIV2L, 170A, DDX13, HLP, SKI2, SKI2W, SKIV2; superki... 62.8 3e-10
tgo:TGME49_085660 DEAD/DEAH box helicase domain-containing pro... 62.4 3e-10
ath:AT3G46960 ATP binding / ATP-dependent helicase/ helicase/ ... 61.6 6e-10
bbo:BBOV_II002350 18.m06191; helicase with zinc finger motif p... 60.5 1e-09
sce:YLR398C SKI2; Ski complex component and putative RNA helic... 57.8 8e-09
tpv:TP04_0364 hypothetical protein; K12599 antiviral helicase ... 57.8 9e-09
cel:F01G4.3 hypothetical protein; K12599 antiviral helicase SK... 54.3 9e-08
cpv:cgd3_280 mRNA translation inhibitor SKI2 SFII helicase, DE... 52.4 4e-07
pfa:PFI0480w helicase with Zn-finger motif, putative; K12599 a... 46.2 2e-05
tgo:TGME49_111900 hypothetical protein 30.0 2.0
ath:AT1G27180 disease resistance protein (TIR-NBS-LRR class), ... 30.0 2.1
tgo:TGME49_041150 hypothetical protein 29.3 3.3
xla:431824 clp1, MGC78822; CLP1, cleavage and polyadenylation ... 29.3 3.5
ath:AT5G06220 hypothetical protein 28.5 6.3
ath:AT1G70070 EMB25; EMB25 (EMBRYO DEFECTIVE 25); ATP-dependen... 27.7 9.6
> tgo:TGME49_013770 RNA helicase, putative ; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1206
Score = 102 bits (255), Expect = 2e-22, Method: Composition-based stats.
Identities = 45/77 (58%), Positives = 65/77 (84%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
+++LTY WA+G+ FA ++ TS+YEGTVIRC+RRLEEL+RQ+A A+K+IGN +LE K +
Sbjct 1130 IMSLTYRWAKGEKFADVLSGTSIYEGTVIRCLRRLEELMRQMACASKSIGNPDLEKKFLE 1189
Query 61 AVRQLRRGSIFSSSLYL 77
++++RRG +FSSSLYL
Sbjct 1190 GIKKIRRGIVFSSSLYL 1206
> cpv:cgd8_2520 Mtr4p like SKI family SFII helicase ; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1280
Score = 96.3 bits (238), Expect = 2e-20, Method: Composition-based stats.
Identities = 41/70 (58%), Positives = 57/70 (81%), Gaps = 0/70 (0%)
Query 8 WAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLRR 67
W++G+SFA+++ +TS YEG+VIRC+RRLEELLRQ+ASA K+IGN +LE K + +RR
Sbjct 1211 WSKGESFASILENTSFYEGSVIRCLRRLEELLRQVASATKSIGNDDLEKKLKEGIALIRR 1270
Query 68 GSIFSSSLYL 77
G +F+ SLYL
Sbjct 1271 GIVFTPSLYL 1280
> dre:406795 skiv2l2, wu:fd11a05, zgc:63838; superkiller viralicidic
activity 2-like 2 (EC:3.6.4.13); K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1034
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 40/77 (51%), Positives = 57/77 (74%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G SF+ + + T ++EG++IRCMRRLEELLRQ+ AAKAIGN ELE K
Sbjct 958 LMDVVYTWANGSSFSQICKMTDVFEGSIIRCMRRLEELLRQMCQAAKAIGNTELENKFAT 1017
Query 61 AVRQLRRGSIFSSSLYL 77
+ +++R +F++SLYL
Sbjct 1018 GITKIKRDIVFAASLYL 1034
> mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viralicidic
activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1040
Score = 92.8 bits (229), Expect = 2e-19, Method: Composition-based stats.
Identities = 40/77 (51%), Positives = 57/77 (74%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G +FA + + T ++EG++IRCMRRLEELLRQ+ AAKAIGN ELE K
Sbjct 964 LMDVVYTWATGATFAHICKMTDVFEGSIIRCMRRLEELLRQMCQAAKAIGNTELENKFAE 1023
Query 61 AVRQLRRGSIFSSSLYL 77
+ +++R +F++SLYL
Sbjct 1024 GITKIKRDIVFAASLYL 1040
> hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118;
superkiller viralicidic activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1042
Score = 92.8 bits (229), Expect = 2e-19, Method: Composition-based stats.
Identities = 40/77 (51%), Positives = 57/77 (74%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G +FA + + T ++EG++IRCMRRLEELLRQ+ AAKAIGN ELE K
Sbjct 966 LMDVVYTWATGATFAHICKMTDVFEGSIIRCMRRLEELLRQMCQAAKAIGNTELENKFAE 1025
Query 61 AVRQLRRGSIFSSSLYL 77
+ +++R +F++SLYL
Sbjct 1026 GITKIKRDIVFAASLYL 1042
> bbo:BBOV_II005660 18.m06470; DSHCT (NUC185) domain containing
DEAD/DEAH box helicase family protein (EC:3.6.1.3); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=986
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 39/77 (50%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
++T+ WA G SFA +++ ++L+EG+VIR +RRLEELLRQLA +++IGN ++E K V
Sbjct 910 MMTVVLKWANGLSFAEVMQESTLFEGSVIRGVRRLEELLRQLACTSRSIGNLQMEQKFVT 969
Query 61 AVRQLRRGSIFSSSLYL 77
+ +L++G IF+SSLYL
Sbjct 970 CINKLKKGIIFTSSLYL 986
> ath:AT2G06990 HEN2; HEN2 (hua enhancer 2); ATP-dependent helicase/
RNA helicase; K12598 ATP-dependent RNA helicase DOB1
[EC:3.6.4.13]
Length=995
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 38/77 (49%), Positives = 56/77 (72%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y W++G SFA +I+ T ++EG++IR RRL+E L QL +AA+A+G LE+K A
Sbjct 919 LMDVIYSWSKGASFAEIIQMTDIFEGSIIRSARRLDEFLNQLRAAAEAVGESSLESKFAA 978
Query 61 AVRQLRRGSIFSSSLYL 77
A LRRG +F++SLYL
Sbjct 979 ASESLRRGIMFANSLYL 995
> cel:W08D2.7 mtr-4; yeast MTR (mRNA TRansport) homolog family
member (mtr-4); K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1026
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 34/77 (44%), Positives = 58/77 (75%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y W G +F+ ++++T ++EG++IR +RRLEE+LR++ +AAKA+ N+ELE K
Sbjct 950 LMDVVYQWVNGATFSEIVKTTDVFEGSIIRTLRRLEEVLREMINAAKALANKELEQKFED 1009
Query 61 AVRQLRRGSIFSSSLYL 77
A + L+R +F++SLYL
Sbjct 1010 ARKNLKRDIVFAASLYL 1026
> ath:AT1G59760 ATP-dependent RNA helicase, putative; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=988
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 33/72 (45%), Positives = 52/72 (72%), Gaps = 0/72 (0%)
Query 6 YGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQL 65
Y WA+G F ++ ++EG++IR +RR+EE+L+QL AAK+IG +LEAK AV ++
Sbjct 917 YAWAKGSKFYEVMEIARVFEGSLIRAIRRMEEVLQQLIVAAKSIGETQLEAKLEEAVSKI 976
Query 66 RRGSIFSSSLYL 77
+R +F++SLYL
Sbjct 977 KRDIVFAASLYL 988
> pfa:PFF0100w ATP-dependent RNA Helicase, putative (EC:3.6.1.-);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1350
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 31/70 (44%), Positives = 45/70 (64%), Gaps = 0/70 (0%)
Query 8 WAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLRR 67
WA+G SF ++ + +YEG++IR +RRL+EL+RQ+ A + I N + A +LRR
Sbjct 1281 WARGHSFVEILSDSQIYEGSIIRTLRRLDELIRQMICAFRGINNDSMCETLTLATNKLRR 1340
Query 68 GSIFSSSLYL 77
G FS SLYL
Sbjct 1341 GIPFSPSLYL 1350
> tpv:TP02_0517 hypothetical protein; K12598 ATP-dependent RNA
helicase DOB1 [EC:3.6.4.13]
Length=1012
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 28/55 (50%), Positives = 43/55 (78%), Gaps = 0/55 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELE 55
L+++ Y WA+G F ++ +S++EG+VIRC+RRL+ELLRQLA A++ IGN +E
Sbjct 935 LMSVVYRWAKGDPFIEILADSSVFEGSVIRCIRRLDELLRQLACASRNIGNMTME 989
> dre:559653 skiv2l, fb70b07, wu:fb70b07; superkiller viralicidic
activity 2 (S. cerevisiae homolog)-like; K12599 antiviral
helicase SKI2 [EC:3.6.4.-]
Length=1230
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 29/76 (38%), Positives = 51/76 (67%), Gaps = 0/76 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L + Y WA+G FA + + T + EGT++RC++RL+E+L+++ AA+ +G+ L +K
Sbjct 1153 LTEVVYCWARGMPFAEIAQLTDVQEGTIVRCIQRLDEVLKEVRQAARIVGDSVLGSKMER 1212
Query 61 AVRQLRRGSIFSSSLY 76
A +RR +F++SLY
Sbjct 1213 ASLAIRRDIVFTASLY 1228
> sce:YJL050W MTR4, DOB1; ATP-dependent 3'-5' RNA helicase, involved
in nuclear RNA processing and degredation both as a component
of the TRAMP complex and in TRAMP independent processes;
member of the Dead-box family of helicases (EC:3.6.1.-);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1073
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y W +G +F + + T +YEG++IR +RLEEL+++L A IGN L+ K A
Sbjct 997 LMEVVYEWCRGATFTQICKMTDVYEGSLIRMFKRLEELVKELVDVANTIGNSSLKEKMEA 1056
Query 61 AVRQLRRGSIFSSSLYL 77
++ + R + + SLYL
Sbjct 1057 VLKLIHRDIVSAGSLYL 1073
> mmu:108077 Skiv2l, 4930534J06Rik, AW214248, Ddx13, SKI, Ski2w;
superkiller viralicidic activity 2-like (S. cerevisiae);
K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1244
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 32/76 (42%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
LV + Y WA+G F+ L + EG V+RC++RL E+ R L AA+ +G L AK
Sbjct 1167 LVEVVYEWARGMPFSELAGLSGTPEGLVVRCIQRLAEMCRSLRGAARLVGEPVLGAKMET 1226
Query 61 AVRQLRRGSIFSSSLY 76
A LRR +F++SLY
Sbjct 1227 AATLLRRDIVFAASLY 1242
> hsa:6499 SKIV2L, 170A, DDX13, HLP, SKI2, SKI2W, SKIV2; superkiller
viralicidic activity 2-like (S. cerevisiae); K12599 antiviral
helicase SKI2 [EC:3.6.4.-]
Length=1246
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 32/76 (42%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
LV + Y WA+G F+ L + EG V+RC++RL E+ R L AA+ +G L AK
Sbjct 1169 LVEVVYEWARGMPFSELAGLSGTPEGLVVRCIQRLAEMCRSLRGAARLVGEPVLGAKMET 1228
Query 61 AVRQLRRGSIFSSSLY 76
A LRR +F++SLY
Sbjct 1229 AATLLRRDIVFAASLY 1244
> tgo:TGME49_085660 DEAD/DEAH box helicase domain-containing protein
; K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1329
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L + Y WA G SF ++ T+ EG+++R + RL+ELLR++ AA IG+ +L AK
Sbjct 1252 LSLVAYDWANGVSFGDIMHKTNAQEGSIVRAILRLDELLRKIRQAAILIGDPDLGAKLQQ 1311
Query 61 AVRQLRRGSIFSSSLYL 77
++RR +F+ SLYL
Sbjct 1312 TSDRIRRDIVFAMSLYL 1328
> ath:AT3G46960 ATP binding / ATP-dependent helicase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1347
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 46/77 (59%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
LV + Y WA+G FA + T + EG ++R + RL+E R+ +AA +GN L K A
Sbjct 1268 LVEVVYEWAKGTPFAEICELTDVPEGLIVRTIVRLDETCREFKNAAAIMGNSALHKKMDA 1327
Query 61 AVRQLRRGSIFSSSLYL 77
A ++R +F++SLY+
Sbjct 1328 ASNAIKRDIVFAASLYV 1344
> bbo:BBOV_II002350 18.m06191; helicase with zinc finger motif
protein; K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1113
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 45/77 (58%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L + Y WA G F+ +++ T L EG ++R + RL+EL R++ A G+Q L++K
Sbjct 1036 LSYVIYQWAIGTPFSEIMQYTDLQEGHIVRAITRLDELCRKIGQVANINGDQALQSKIEK 1095
Query 61 AVRQLRRGSIFSSSLYL 77
++RG +F SLYL
Sbjct 1096 VSNSIKRGIVFMPSLYL 1112
> sce:YLR398C SKI2; Ski complex component and putative RNA helicase,
mediates 3'-5' RNA degradation by the cytoplasmic exosome;
null mutants have superkiller phenotype of increased viral
dsRNAs and are synthetic lethal with mutations in 5'-3'
mRNA decay (EC:3.6.1.-); K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1287
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
++ + Y WA+G SF ++ + EGTV+R + L+E+ R++ +A+ IGN L K
Sbjct 1211 MMNVVYEWARGLSFKEIMEMSPEAEGTVVRVITWLDEICREVKTASIIIGNSTLHMKMSR 1270
Query 61 AVRQLRRGSIFSSSLYL 77
A ++R +F++SLYL
Sbjct 1271 AQELIKRDIVFAASLYL 1287
> tpv:TP04_0364 hypothetical protein; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1069
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L + Y WA G F ++ T L EG ++R + RL+EL R++ A G+Q+L K
Sbjct 992 LSYVIYQWASGTPFQEIMELTDLQEGHIVRVILRLDELCRKILQTANIFGHQKLAEKIEL 1051
Query 61 AVRQLRRGSIFSSSLYL 77
+RR +F SLYL
Sbjct 1052 VCNAIRRDIVFKQSLYL 1068
> cel:F01G4.3 hypothetical protein; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1266
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 44/76 (57%), Gaps = 0/76 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
L+ + Y WA G F ++ T EG +++C++RL+E+ + + +A + +G+ L K
Sbjct 1188 LMEVVYEWANGTPFYQIMEMTDCQEGLIVKCIQRLDEVCKDVRNAGRIVGDPALVEKMEE 1247
Query 61 AVRQLRRGSIFSSSLY 76
+RR +F++SLY
Sbjct 1248 VSASIRRDIVFAASLY 1263
> cpv:cgd3_280 mRNA translation inhibitor SKI2 SFII helicase,
DEXDc+HELICc ; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1439
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 5/79 (6%)
Query 4 LTYGWAQGQSFAALIR-----STSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKC 58
+ Y W+ +S ++ +L+EGT++R + RL+EL+R+L AAK +G++ LE K
Sbjct 1358 IAYKWSNKESLKEIMEFVHNSEMNLHEGTIVRTILRLDELVRKLIIAAKMMGDKILEEKL 1417
Query 59 VAAVRQLRRGSIFSSSLYL 77
+ R IF +SLY
Sbjct 1418 CLIHENIARDIIFMTSLYF 1436
> pfa:PFI0480w helicase with Zn-finger motif, putative; K12599
antiviral helicase SKI2 [EC:3.6.4.-]
Length=1373
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 44/77 (57%), Gaps = 0/77 (0%)
Query 1 LVTLTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVA 60
++ + Y W G SFA L+ L EG ++R + RL++L R++ A +GN +L K
Sbjct 1296 IMFIAYKWTLGVSFAELLEQCELEEGLIVRSILRLDDLCRKVKIAFLYLGNIDLAQKVEK 1355
Query 61 AVRQLRRGSIFSSSLYL 77
LRR IF++SLYL
Sbjct 1356 TSHLLRRDIIFTTSLYL 1372
> tgo:TGME49_111900 hypothetical protein
Length=340
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 22 SLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAKC 58
+L TV+ +R++ E RQL AAK G +E A C
Sbjct 76 NLVRSTVVSHLRKVAETSRQLEEAAKEEGQRETTAAC 112
> ath:AT1G27180 disease resistance protein (TIR-NBS-LRR class),
putative
Length=1556
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 7/58 (12%)
Query 16 ALIRSTSLY-EGTVIRCMRRLEELLRQLASAAKAIGNQELEAKCVAAVRQLRRGSIFS 72
AL+ TS Y EG++I R E+L +L+ NQ+ E KC+ + L+ S +S
Sbjct 484 ALVGETSWYGEGSLIVITTRDSEILSKLSV------NQQYEVKCLTEPQALKLFSFYS 535
> tgo:TGME49_041150 hypothetical protein
Length=592
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 9 AQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLASAAKAIGNQELEAK 57
A+ + FA++ ++ LYE + + R+E LR+LA KA+ + ++++
Sbjct 54 AELEHFASVRQAFELYEQDALMEVHRIERHLRRLAPEDKALLQESIDSR 102
> xla:431824 clp1, MGC78822; CLP1, cleavage and polyadenylation
factor I subunit, homolog (EC:2.7.1.78); K14399 polyribonucleotide
5'-hydroxyl-kinase [EC:2.7.1.78]
Length=439
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 35/84 (41%), Gaps = 17/84 (20%)
Query 5 TYGWAQGQSFAALIRSTSLYEGTVIRCM---RRLEELLRQLASAAKAI------GNQELE 55
T GW +G + ALI + S +E V+ + R +LLR L + + G E
Sbjct 242 TCGWVKGSGYQALIHAASAFEVDVVLVLDQERLYNDLLRDLPHFVRTLLLPKSGGASERS 301
Query 56 AKCVAAVRQLR--------RGSIF 71
+C R R RGS++
Sbjct 302 KECRRESRDQRVREYFYGPRGSLY 325
> ath:AT5G06220 hypothetical protein
Length=813
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 28/40 (70%), Gaps = 2/40 (5%)
Query 34 RLEELLRQLASAAKAIGNQELEAKC--VAAVRQLRRGSIF 71
RLE LL+QL +++ + G ++++A C + +R+L++ + F
Sbjct 501 RLESLLQQLHASSSSSGKEQIKAACSDLEKIRKLKKEAEF 540
> ath:AT1G70070 EMB25; EMB25 (EMBRYO DEFECTIVE 25); ATP-dependent
helicase/ RNA helicase
Length=1171
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 4 LTYGWAQGQSFAALIRSTSLYEGTVIRCMRRLEELLRQLA 43
+ WA G S+ ++ ++ EG + R +RR +LL Q+
Sbjct 1102 MVEAWASGLSWKEMMMECAMDEGDLARLLRRTIDLLAQIP 1141
Lambda K H
0.323 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072310352
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40