bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2823_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
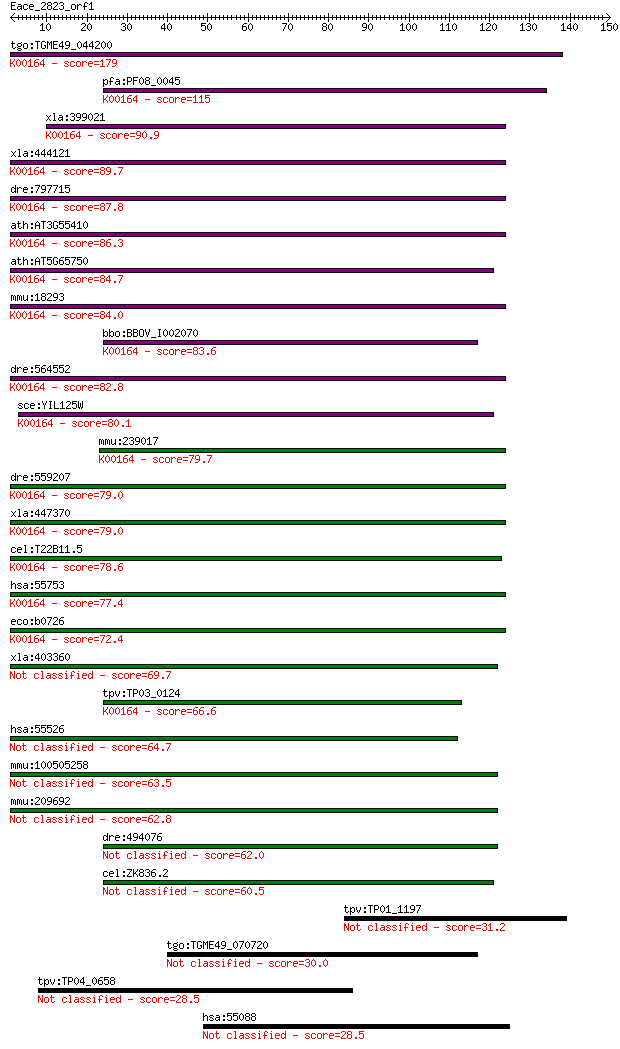
tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1... 179 3e-45
pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.... 115 6e-26
xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alp... 90.9 1e-18
xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogena... 89.7 3e-18
dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogen... 87.8 1e-17
ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putat... 86.3 3e-17
ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putat... 84.7 1e-16
mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA41... 84.0 1e-16
bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1 co... 83.6 2e-16
dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,... 82.8 3e-16
sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-k... 80.1 3e-15
mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-)... 79.7 3e-15
dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehyd... 79.0 5e-15
xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like; K... 79.0 6e-15
cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehyd... 78.6 7e-15
hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);... 77.4 2e-14
eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxy... 72.4 5e-13
xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolas... 69.7 3e-12
tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.... 66.6 3e-11
hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogena... 64.7 1e-10
mmu:100505258 probable 2-oxoglutarate dehydrogenase E1 compone... 63.5 2e-10
mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transke... 62.8 4e-10
dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketol... 62.0 8e-10
cel:ZK836.2 hypothetical protein 60.5 2e-09
tpv:TP01_1197 hypothetical protein 31.2 1.3
tgo:TGME49_070720 hypothetical protein 30.0 2.5
tpv:TP04_0658 hypothetical protein 28.5 8.5
hsa:55088 C10orf118, FLJ10188, FLJ35301, MGC118918, MGC129699;... 28.5 8.8
> tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1116
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 86/137 (62%), Positives = 107/137 (78%), Gaps = 2/137 (1%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YDL A ++++++ D G ++AIAR+EQLSPFPFDL I+D++R+PNL+SVVWAQEEP
Sbjct 979 YDLIAGKDKMKNGDENGD--GDKIAIARMEQLSPFPFDLFIEDLKRFPNLKSVVWAQEEP 1036
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
MNQGAW YTSKRIES L HL PN I P+Y GRDV A TAVGDK LHD+ELAQLL DA
Sbjct 1037 MNQGAWFYTSKRIESSLRHLNFPNGIRSPIYAGRDVCAATAVGDKKLHDQELAQLLQDAL 1096
Query 121 NLECTDNSYLSKYLSRR 137
++ T +SYL KYL ++
Sbjct 1097 DINRTTHSYLEKYLHKQ 1113
> pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1038
Score = 115 bits (287), Expect = 6e-26, Method: Composition-based stats.
Identities = 57/115 (49%), Positives = 76/115 (66%), Gaps = 5/115 (4%)
Query 24 VAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIESCLFHLGRP 83
VAIAR+EQLSPFPF ++ D+Q YPNLR ++WAQEE MN G W Y S+RIE+ + L +
Sbjct 920 VAIARIEQLSPFPFKQIMNDLQTYPNLRDIIWAQEEHMNMGPWFYVSRRIEASIKQLKKD 979
Query 84 N-----KIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAFNLECTDNSYLSKY 133
+I + Y GRDV A + GD +LH +L + LVDAFNL+ N ++ KY
Sbjct 980 KPKWNIQIPQVRYSGRDVYAAQSAGDLNLHLYQLDEFLVDAFNLDKKYNMHVQKY 1034
> xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1021
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 55/114 (48%), Positives = 68/114 (59%), Gaps = 8/114 (7%)
Query 10 LQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYT 69
L E S D G VAIARVEQLSPFPFDLV K++Q+YPN +VW QEE NQG + Y
Sbjct 910 LTKERSGRDMEG-DVAIARVEQLSPFPFDLVEKEVQKYPN-ADLVWCQEEHKNQGYYDYV 967
Query 70 SKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAFNLE 123
R+ + + H +P Y GRD +A A G+K H EL + L AFNL+
Sbjct 968 KPRLRTTI-HRTKP-----VWYAGRDPAAAPATGNKKTHLTELRRFLDTAFNLD 1015
> xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1018
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 55/123 (44%), Positives = 69/123 (56%), Gaps = 15/123 (12%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YDL ER E VAI RVEQLSPFPFDLV K++Q+YPN +VW QEE
Sbjct 905 YDLTKERSGRGMEGD--------VAITRVEQLSPFPFDLVEKEVQKYPN-ADLVWCQEEH 955
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
NQG + Y R+ + + H +P Y GRD +A A G+K H EL +LL +F
Sbjct 956 KNQGYYDYVKPRLRTTI-HRAKPV-----WYAGRDPAAAPATGNKKTHLTELKRLLDTSF 1009
Query 121 NLE 123
NL+
Sbjct 1010 NLD 1012
> dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1023
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 54/123 (43%), Positives = 68/123 (55%), Gaps = 15/123 (12%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YDL ER+ E RVAI+R+EQLSPFPFDLV + ++YP+ ++W QEE
Sbjct 910 YDLQRERKSRGLEE--------RVAISRIEQLSPFPFDLVKAEAEKYPHAH-LLWCQEEH 960
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
NQG + Y RI + L N YVGRD +A A G+K H EL + L AF
Sbjct 961 KNQGYYDYVKPRISTTL------NNTRPVWYVGRDPAAAPATGNKKAHLLELQRFLDTAF 1014
Query 121 NLE 123
NLE
Sbjct 1015 NLE 1017
> ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1017
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 53/123 (43%), Positives = 72/123 (58%), Gaps = 11/123 (8%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y+L ER+++ A D VAI RVEQL PFP+DL+ ++++RYPN +VW QEE
Sbjct 903 YELDDERKKV----GATD-----VAICRVEQLCPFPYDLIQRELKRYPNAE-IVWCQEEA 952
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
MN GA+ Y S R+ + + + R + +E YVGR SA TA G H KE A L+ A
Sbjct 953 MNMGAFSYISPRLWTAMRSVNRGD-MEDIKYVGRGPSAATATGFYTFHVKEQAGLVQKAI 1011
Query 121 NLE 123
E
Sbjct 1012 GKE 1014
> ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1025
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 50/120 (41%), Positives = 69/120 (57%), Gaps = 11/120 (9%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y+L ER++ +++ VAI RVEQL PFP+DL+ ++++RYPN +VW QEEP
Sbjct 907 YELDEERKKSETKD---------VAICRVEQLCPFPYDLIQRELKRYPNAE-IVWCQEEP 956
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
MN G + Y + R+ + + L R N YVGR SA TA G LH KE L+ A
Sbjct 957 MNMGGYQYIALRLCTAMKALQRGN-FNDIKYVGRLPSAATATGFYQLHVKEQTDLVKKAL 1015
> mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA4192,
d1401, mKIAA4192; oxoglutarate dehydrogenase (lipoamide)
(EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1023
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 50/123 (40%), Positives = 69/123 (56%), Gaps = 15/123 (12%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YDL ER+ A N VAI R+EQLSPFPFDL++K+ Q+YPN + W QEE
Sbjct 910 YDLTRERK--------ARNMEEEVAITRIEQLSPFPFDLLLKEAQKYPN-AELAWCQEEH 960
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
NQG + Y R+ + + ++ + Y GRD +A A G+K H EL + L AF
Sbjct 961 KNQGYYDYVKPRLRTTI------DRAKPVWYAGRDPAAAPATGNKKTHLTELQRFLDTAF 1014
Query 121 NLE 123
+L+
Sbjct 1015 DLD 1017
> bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1
component (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=891
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 42/93 (45%), Positives = 54/93 (58%), Gaps = 3/93 (3%)
Query 24 VAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIESCLFHLGRP 83
VA+ VEQL PFP + +++RYPNL+ +VW QEE N G W Y S RI S L HLG
Sbjct 795 VAVTTVEQLCPFPAGALKSELERYPNLKRLVWCQEEHANAGGWSYVSPRICSLLAHLGSG 854
Query 84 NKIERPLYVGRDVSATTAVGDKHLHDKELAQLL 116
++E YVGR + + GD H EL + L
Sbjct 855 LRLE---YVGRPPLSAPSCGDSRTHGVELQKFL 884
> dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,
zgc:73296; oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1022
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 49/123 (39%), Positives = 70/123 (56%), Gaps = 15/123 (12%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y+L ER+ E+S VAI R+EQLSPFPFDLV + +++PN +VW QEE
Sbjct 909 YELTRERKARNMENS--------VAITRIEQLSPFPFDLVRAETEKFPN-ADLVWCQEEH 959
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
NQG + Y R+ + + N+ + Y GR+ +A A G+K+ H EL + L AF
Sbjct 960 KNQGYYDYVKPRMRTTI------NRTKPVWYAGREPAAAPATGNKNTHLLELKRFLDTAF 1013
Query 121 NLE 123
NL+
Sbjct 1014 NLD 1016
> sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-ketoglutarate
dehydrogenase complex, which catalyzes a key
step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation
of alpha-ketoglutarate to form succinyl-CoA (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1014
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 44/118 (37%), Positives = 59/118 (50%), Gaps = 10/118 (8%)
Query 3 LHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMN 62
LH RE L +++A ++EQL PFPF + + YPNL +VW QEEP+N
Sbjct 904 LHKRRESLGDKTTA---------FLKIEQLHPFPFAQLRDSLNSYPNLEEIVWCQEEPLN 954
Query 63 QGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
G+W YT R+ + L + K + Y GR+ S A G K LH E L D F
Sbjct 955 MGSWAYTEPRLHTTLKETDK-YKDFKVRYCGRNPSGAVAAGSKSLHLAEEDAFLKDVF 1011
> mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1029
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 44/102 (43%), Positives = 62/102 (60%), Gaps = 9/102 (8%)
Query 23 RVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIESCLFHLGR 82
+VAI R+EQ+SPFPFDL++++ ++Y +VW QEE N G + Y S R + L H
Sbjct 929 QVAITRLEQISPFPFDLIMREAEKYSGAE-LVWCQEEHKNMGYYDYISPRFMTLLGH--- 984
Query 83 PNKIERPL-YVGRDVSATTAVGDKHLHDKELAQLLVDAFNLE 123
RP+ YVGRD +A A G+K+ H L + L AFNL+
Sbjct 985 ----SRPIWYVGRDPAAAPATGNKNAHLVSLRRFLDTAFNLK 1022
> dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1008
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 50/124 (40%), Positives = 71/124 (57%), Gaps = 18/124 (14%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y+L ER++L+ E + VAI R+EQ+SPFPFDL+ ++++Y N ++W QEE
Sbjct 895 YELAKERKQLKLEEN--------VAIVRLEQISPFPFDLIKAEVEKYSN-AELIWCQEEH 945
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPL-YVGRDVSATTAVGDKHLHDKELAQLLVDA 119
N G + Y R F +P + P+ YVGRD +A A G+K H EL + L A
Sbjct 946 KNMGYYDYIRPR-----FLTVQP---KNPIWYVGRDPAAAPATGNKFTHLAELKRFLDTA 997
Query 120 FNLE 123
FNLE
Sbjct 998 FNLE 1001
> xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like;
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1018
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 51/124 (41%), Positives = 69/124 (55%), Gaps = 17/124 (13%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y+L ER R +S +VAI R+EQ+SPFPFDLV ++ ++Y +VW QEE
Sbjct 904 YELVKERHRKGLDS--------QVAITRLEQISPFPFDLVKQEAEKYAT-SELVWCQEEH 954
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPL-YVGRDVSATTAVGDKHLHDKELAQLLVDA 119
N G + Y R + L H RP+ YVGRD +A A G+K+ H EL + L A
Sbjct 955 KNMGYYDYVKARFLTILNH-------ARPVWYVGRDPAAAPATGNKNTHHVELRRFLDIA 1007
Query 120 FNLE 123
F+LE
Sbjct 1008 FDLE 1011
> cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1029
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 46/122 (37%), Positives = 67/122 (54%), Gaps = 14/122 (11%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YD+ A R+ + E+ VA+ RVEQLSPFP+DLV ++ ++Y ++WAQEE
Sbjct 912 YDMVAARKHVGKEND--------VALVRVEQLSPFPYDLVQQECRKYQGAE-ILWAQEEH 962
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
N GAW + RI S L GR K Y GR S++ A G+K H +E +++ F
Sbjct 963 KNMGAWSFVQPRINSLLSIDGRATK-----YAGRLPSSSPATGNKFTHMQEQKEMMSKVF 1017
Query 121 NL 122
+
Sbjct 1018 GV 1019
> hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=953
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 48/124 (38%), Positives = 67/124 (54%), Gaps = 17/124 (13%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YDL ER E +VAI R+EQ+SPFPFDL+ ++ ++YP + W QEE
Sbjct 839 YDLVKERSSQDLEE--------KVAITRLEQISPFPFDLIKQEAEKYPG-AELAWCQEEH 889
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPL-YVGRDVSATTAVGDKHLHDKELAQLLVDA 119
N G + Y S R + L + RP+ YVGRD +A A G+++ H L + L A
Sbjct 890 KNMGYYDYISPRFMTIL-------RRARPIWYVGRDPAAAPATGNRNTHLVSLKKFLDTA 942
Query 120 FNLE 123
FNL+
Sbjct 943 FNLQ 946
> eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxylase,
thiamin-requiring (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=933
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 66/123 (53%), Gaps = 15/123 (12%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
YDL +R + N + VAI R+EQL PFP + + +Q++ +++ VW QEEP
Sbjct 826 YDLLEQRRK---------NNQHDVAIVRIEQLYPFPHKAMQEVLQQFAHVKDFVWCQEEP 876
Query 61 MNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
+NQGAW Y S+ + G + Y GR SA+ AVG +H K+ L+ DA
Sbjct 877 LNQGAW-YCSQHHFREVIPFGASLR-----YAGRPASASPAVGYMSVHQKQQQDLVNDAL 930
Query 121 NLE 123
N+E
Sbjct 931 NVE 933
> xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=927
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 59/124 (47%), Gaps = 21/124 (16%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y LH +RE L + G AI RVE+L PFP + + ++I RYP + +W+QEEP
Sbjct 822 YALHKQREALGEQ-------GRSSAIIRVEELCPFPLEALQQEIHRYPKAKDFIWSQEEP 874
Query 61 MNQGAWGYTSKRIE---SCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLV 117
N GAW + + R E +C L V R AVG LH ++ ++ V
Sbjct 875 QNMGAWTFVAPRFEKQLACKLRL-----------VSRPALPAPAVGIGTLHQQQQEEITV 923
Query 118 DAFN 121
+
Sbjct 924 KTLS 927
> tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1030
Score = 66.6 bits (161), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/93 (35%), Positives = 57/93 (61%), Gaps = 4/93 (4%)
Query 24 VAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIESCLFHL--- 80
+ +AR+E+++PFP ++ D++++ NL ++VW QEE N G + + +R+ + L L
Sbjct 929 IPVARIEEITPFPAQNILDDLKKFKNLETLVWCQEEHENSGCFYFLRERLNNVLKILKNE 988
Query 81 GRPNKIERPL-YVGRDVSATTAVGDKHLHDKEL 112
G K+ + Y GR ATTAVGD +H+ E+
Sbjct 989 GHCPKVNTAVKYSGRYPCATTAVGDPKMHEHEI 1021
> hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogenase
E1 and transketolase domain containing 1 (EC:1.2.4.2)
Length=919
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 58/115 (50%), Gaps = 21/115 (18%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y L +RE L ++ + AI RVE+L PFP D + +++ +Y +++ +W+QEEP
Sbjct 814 YSLVKQRESLGAKK-------HDFAIIRVEELCPFPLDSLQQEMSKYKHVKDHIWSQEEP 866
Query 61 MNQGAWGYTSKRIE---SCLFHL-GRPNKIERPLYVGRDVSATTAVGDKHLHDKE 111
N G W + S R E +C L GRP PL V +G HLH E
Sbjct 867 QNMGPWSFVSPRFEKQLACKLRLVGRP-----PLPV-----PAVGIGTVHLHQHE 911
> mmu:100505258 probable 2-oxoglutarate dehydrogenase E1 component
DHKTD1, mitochondrial-like
Length=532
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 59/124 (47%), Gaps = 21/124 (16%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y L +RE L ++ + AI R+E+L PFP D + +++ +Y ++R V+W+QEEP
Sbjct 426 YALLKQRESLGTKK-------HDFAIIRLEELCPFPLDALQQEMSKYKHVRDVIWSQEEP 478
Query 61 MNQGAWGYTSKRIE---SCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLV 117
N G W + S R E +C L V R AVG +H ++ +L
Sbjct 479 QNMGPWSFVSPRFEKQLACRLRL-----------VSRPPLPAPAVGIGTVHQQQHEDILS 527
Query 118 DAFN 121
F
Sbjct 528 KTFT 531
> mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=921
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 59/124 (47%), Gaps = 21/124 (16%)
Query 1 YDLHAERERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEP 60
Y L +RE L ++ + AI R+E+L PFP D + +++ +Y ++R V+W+QEEP
Sbjct 815 YALLKQRESLGTKK-------HDFAIIRLEELCPFPLDALQQEMSKYKHVRDVIWSQEEP 867
Query 61 MNQGAWGYTSKRIE---SCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLV 117
N G W + S R E +C L V R AVG +H ++ +L
Sbjct 868 QNMGPWSFVSPRFEKQLACRLRL-----------VSRPPLPAPAVGIGTVHQQQHEDILS 916
Query 118 DAFN 121
F
Sbjct 917 KTFT 920
> dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=925
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 50/101 (49%), Gaps = 14/101 (13%)
Query 24 VAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIE---SCLFHL 80
A+ RVE+L PFP + + +++ +Y N + +W+QEEP N G W + S R E +C L
Sbjct 836 TALVRVEELCPFPTEALQQELNKYTNAKEFIWSQEEPQNMGCWSFVSPRFEKQLACKLRL 895
Query 81 GRPNKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAFN 121
V R AVG LH ++ +L +F+
Sbjct 896 -----------VSRPALPAPAVGIGTLHHQQHEAILTSSFS 925
> cel:ZK836.2 hypothetical protein
Length=911
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 47/97 (48%), Gaps = 8/97 (8%)
Query 24 VAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIESCLFHLGRP 83
VAI RVE L PFP + +++YP + VW+QEEP N GAW + R E+ L
Sbjct 822 VAIVRVEMLCPFPVVDLQAVLKKYPGAQDFVWSQEEPRNAGAWSFVRPRFENALG----- 876
Query 84 NKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAF 120
R + GR A TA H KE +++ F
Sbjct 877 ---VRLKFAGRPELAWTATAIGEHHTKEAEEVINQTF 910
> tpv:TP01_1197 hypothetical protein
Length=286
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 5/55 (9%)
Query 84 NKIERPLYVGRDVSATTAVGDKHLHDKELAQLLVDAFNLECTDNSYLSKYLSRRV 138
N + PL ++ TTA L D EL++ ++D+F L SY S Y+ RR+
Sbjct 5 NLLILPLTALISLTGTTA-----LTDVELSERIIDSFKLMYKQISYASYYMKRRI 54
> tgo:TGME49_070720 hypothetical protein
Length=897
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 2/79 (2%)
Query 40 VIKDIQRYPNLRSVVWAQEEPMNQGAWGYTSKRIESCLFHLGRP--NKIERPLYVGRDVS 97
V D QR LR + A P QG G++S+ E+ + +GR ++ER + +
Sbjct 581 VSADKQRLAALRRLGEAALPPETQGVAGFSSRESEALVARVGRAWRMRLERLGGLPAGET 640
Query 98 ATTAVGDKHLHDKELAQLL 116
A +H + LA+LL
Sbjct 641 DAHAAARRHQRGESLAELL 659
> tpv:TP04_0658 hypothetical protein
Length=713
Score = 28.5 bits (62), Expect = 8.5, Method: Composition-based stats.
Identities = 17/78 (21%), Positives = 38/78 (48%), Gaps = 3/78 (3%)
Query 8 ERLQSESSAADNPGYRVAIARVEQLSPFPFDLVIKDIQRYPNLRSVVWAQEEPMNQGAWG 67
+R+QS+ A P V + + + FP+DL+I I+ + + + E + +
Sbjct 194 DRIQSDPFA---PPSNVRVRIPQDTARFPYDLMIPKIRNIASCDYIARSISEELTAMSRE 250
Query 68 YTSKRIESCLFHLGRPNK 85
+ S+ ++ FH+ +P +
Sbjct 251 WESRSLKKRGFHISKPTQ 268
> hsa:55088 C10orf118, FLJ10188, FLJ35301, MGC118918, MGC129699;
chromosome 10 open reading frame 118
Length=898
Score = 28.5 bits (62), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 39/76 (51%), Gaps = 6/76 (7%)
Query 49 NLRSVVWAQEEPMNQGAWGYTSKRIESCLFHLGRPNKIERPLYVGRDVSATTAVGDKHLH 108
NLR + + E Q T K++ES + L + K R + +DV+A AV + LH
Sbjct 232 NLREELKKRTETEKQHM--NTIKQLESRIEELNKEVKASRDQLIAQDVTAKNAV--QQLH 287
Query 109 DKELAQLLVDAFNLEC 124
KE+AQ + A N +C
Sbjct 288 -KEMAQRMEQA-NKKC 301
Lambda K H
0.316 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40