bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2834_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
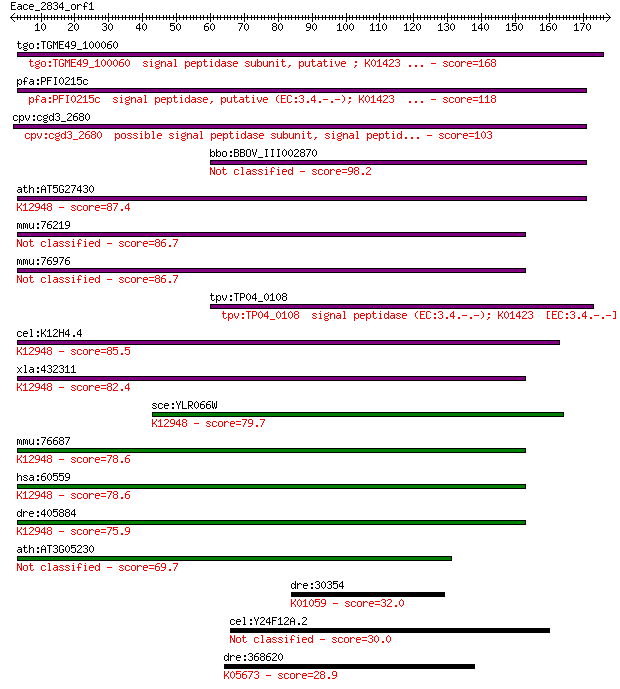
tgo:TGME49_100060 signal peptidase subunit, putative ; K01423 ... 168 8e-42
pfa:PFI0215c signal peptidase, putative (EC:3.4.-.-); K01423 ... 118 1e-26
cpv:cgd3_2680 possible signal peptidase subunit, signal peptid... 103 2e-22
bbo:BBOV_III002870 17.m07272; signal peptidase family protein 98.2 1e-20
ath:AT5G27430 signal peptidase subunit family protein; K12948 ... 87.4 2e-17
mmu:76219 Arxes1, 6530401D17Rik, Spcs3; adipocyte-related X-ch... 86.7 3e-17
mmu:76976 Arxes2, 2900062L11Rik, Spcs3; adipocyte-related X-ch... 86.7 3e-17
tpv:TP04_0108 signal peptidase (EC:3.4.-.-); K01423 [EC:3.4.-.-] 86.3 5e-17
cel:K12H4.4 hypothetical protein; K12948 signal peptidase comp... 85.5 9e-17
xla:432311 spcs3, MGC79052; signal peptidase complex subunit 3... 82.4 8e-16
sce:YLR066W SPC3; Spc3p; K12948 signal peptidase complex subun... 79.7 4e-15
mmu:76687 Spcs3, 1810011E08Rik; signal peptidase complex subun... 78.6 1e-14
hsa:60559 SPCS3, DKFZp564J1864, FLJ22649, PRO3567, SPC22/23, S... 78.6 1e-14
dre:405884 spcs3, MGC85675, zgc:85675; signal peptidase comple... 75.9 7e-14
ath:AT3G05230 signal peptidase subunit family protein 69.7 5e-12
dre:30354 lpl, fb62e04, fc49b03, wu:fb62e04, wu:fc49b03; lipop... 32.0 1.2
cel:Y24F12A.2 hypothetical protein 30.0 4.4
dre:368620 abcc4, MRP, cb1019, si:busm1-52i16.4, si:dz202l16.5... 28.9 9.2
> tgo:TGME49_100060 signal peptidase subunit, putative ; K01423
[EC:3.4.-.-]
Length=175
Score = 168 bits (426), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 81/173 (46%), Positives = 119/173 (68%), Gaps = 0/173 (0%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFFLVGPISGSVSAREVYGFGYNYALSGDQA 62
M+ YLNR NAV C+L+ +L + A+GN S++ +G VS EVY FG N AL G+QA
Sbjct 1 MDTYLNRGNAVVCTLLAALALAAVGNHFSTYLFQADPTGKVSIAEVYEFGVNNALQGEQA 60
Query 63 VLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLANVP 122
++ +I+ADL F WN +QLFV+V+ YE+P++P N+V+V+DRIIT+ +A++D V
Sbjct 61 QVALNIQADLTSCFNWNTQQLFVYVIVRYETPKNPRNEVIVWDRIITDPDDAIIDFEGVI 120
Query 123 AKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQYDRISQ 175
KY LRD G+ LR R +T+ L+ YHP+VG I + VASST+ +P+ Y R ++
Sbjct 121 NKYPLRDNGRSLRNRTVTVALEYAYHPVVGVIKSGHVASSTYTLPSSYFRYAK 173
> pfa:PFI0215c signal peptidase, putative (EC:3.4.-.-); K01423
[EC:3.4.-.-]
Length=185
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 60/170 (35%), Positives = 98/170 (57%), Gaps = 2/170 (1%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFFLVGP--ISGSVSAREVYGFGYNYALSGD 60
M+++LNR N +F S+ + +L N +SF+L I ++ + + F YN ++ D
Sbjct 1 MDSFLNRLNVLFYSMALCFLILCAFNYGTSFYLFDEKEIKTNIQVKSIKRFVYNRYINAD 60
Query 61 QAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLAN 120
+AVLS D+ D+R F WN KQLFV+V+ YE+P+ N+V++ D I+ N+ +A + N
Sbjct 61 EAVLSLDVSYDMRKAFNWNLKQLFVYVLVTYETPKKIKNEVIIQDYIVKNKKQAKKNYRN 120
Query 121 VPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQY 170
KY L+D GLR I L++ Y PIVG + A ++++P +Y
Sbjct 121 FITKYSLKDYYNGLRNNLIHLQVCYKYMPIVGFSRSFEGAKISYQLPPEY 170
> cpv:cgd3_2680 possible signal peptidase subunit, signal peptide
; K01423 [EC:3.4.-.-]
Length=203
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 56/171 (32%), Positives = 87/171 (50%), Gaps = 3/171 (1%)
Query 2 KMENYLNRANAVFCSLMVSLGVLALGNIASSFFLVGPISGSVSAREVYGFGYNYALSGDQ 61
KM++ +R N +FCS ++SL A+GN ASSF G + G + L DQ
Sbjct 30 KMDSLFSRINIIFCSFVISLACCAVGNFASSFIYKELPIGDTELHSILDLGISPYLRNDQ 89
Query 62 AVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIIT-NESEAVLDLAN 120
A ++ +I +L WN Q+F F+ Y++ +H N V V+D I + +++ +
Sbjct 90 ANIALNINTNLSNSLNWNTNQIFTFIYVSYKN-KHQNNYVTVWDDIFSKKKNKTSFSMKG 148
Query 121 VPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRI-YTQTVASSTFRMPAQY 170
V KY +RD G+ LR + I L + Y PIVG I Y S+ ++P Y
Sbjct 149 VINKYPIRDIGRNLRSKSINLNIAFCYMPIVGSIKYHHLKVSTEKKLPVNY 199
> bbo:BBOV_III002870 17.m07272; signal peptidase family protein
Length=171
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 41/111 (36%), Positives = 70/111 (63%), Gaps = 0/111 (0%)
Query 60 DQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLA 119
D+A ++ DLR +F W+A +F++ YE+P+HP N++++FD+IIT++ EA A
Sbjct 59 DRAAFELNLSYDLRDVFDWSANVIFLYATVNYETPKHPVNELIIFDKIITSKEEAYEPGA 118
Query 120 NVPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQY 170
++ +KY++ D + LR +TL+L + PI G I + +A S F MP+ Y
Sbjct 119 DIVSKYYMIDYARSLRKARVTLRLHYCFVPIGGLIKSYQLAESVFTMPSDY 169
> ath:AT5G27430 signal peptidase subunit family protein; K12948
signal peptidase complex subunit 3 [EC:3.4.-.-]
Length=167
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 52/169 (30%), Positives = 85/169 (50%), Gaps = 4/169 (2%)
Query 3 MENYLNRANAVFCSLMVSLG-VLALGNIASSFFLVGPISGSVSAREVYGFGYNYALSGDQ 61
M ++ RANA+ + L + A+ + + +F P S + + F D+
Sbjct 1 MHSFGYRANALLTFAVTILAFICAIASFSDNFSNQNP-SAQIQILNINWFQ-KQPHGNDE 58
Query 62 AVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLANV 121
L+ +I ADL+ LF WN KQ+F FV AEYE+ ++ NQV ++D II + A + +
Sbjct 59 VSLTLNITADLQSLFTWNTKQVFAFVAAEYETSKNALNQVSLWDAIIPEKEHAKFWI-QI 117
Query 122 PAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQY 170
KY D+G LRG++ L L P G+++ + S +R+P Y
Sbjct 118 SNKYRFIDQGHNLRGKDFNLTLHWHVMPKTGKMFADKIVMSGYRLPNAY 166
> mmu:76219 Arxes1, 6530401D17Rik, Spcs3; adipocyte-related X-chromosome
expressed sequence 1
Length=180
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 56/156 (35%), Positives = 83/156 (53%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVS---AREVYGFGYNYAL 57
M + L+RAN++F + + L LG I ++ F P+ VS ++V F
Sbjct 1 MNSLLSRANSLFAFTLSVMAALTLGCILTTAFKDRSAPVRLHVSRILLKKVEDFTGPRKK 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++F I ADL F WN KQLF+++ AEY + + NQVV++D+I+ L+
Sbjct 61 S-DLGFITFHISADLEKTFDWNVKQLFLYLSAEYSTKSNAVNQVVLWDKILLRGENPKLN 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L +V +KY D G GL+G R +TL L PI G
Sbjct 120 LKDVKSKYFFFDDGHGLKGNRNVTLTLSWQVIPIAG 155
> mmu:76976 Arxes2, 2900062L11Rik, Spcs3; adipocyte-related X-chromosome
expressed sequence 2
Length=180
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 56/156 (35%), Positives = 83/156 (53%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVS---AREVYGFGYNYAL 57
M + L+RAN++F + + L LG I ++ F P+ VS ++V F
Sbjct 1 MNSLLSRANSLFAFTLSVMAALTLGCILTTAFKDRSAPVRLHVSRILLKKVEDFTGPRKK 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++F I ADL F WN KQLF+++ AEY + + NQVV++D+I+ L+
Sbjct 61 S-DLGFITFHISADLEKTFDWNVKQLFLYLSAEYSTKSNAVNQVVLWDKILLRGENPKLN 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L +V +KY D G GL+G R +TL L PI G
Sbjct 120 LKDVKSKYFFFDDGHGLKGNRNVTLTLSWQVIPIAG 155
> tpv:TP04_0108 signal peptidase (EC:3.4.-.-); K01423 [EC:3.4.-.-]
Length=155
Score = 86.3 bits (212), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 69/113 (61%), Gaps = 1/113 (0%)
Query 60 DQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLA 119
D+A+L + DLRG+F W+ +F++V A Y + +H ++VV+FD+II N+SEA
Sbjct 43 DRALLELSMGYDLRGVFDWSTHVVFLYVTANYVTNRHERSEVVIFDKII-NKSEAYQPST 101
Query 120 NVPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQYDR 172
NV AKY L D G+ LR R+++LK PI G I ++ TF +P QY +
Sbjct 102 NVFAKYFLYDFGRSLRNRQVSLKFFYEIVPIGGFIKQFQLSHHTFTLPPQYSQ 154
> cel:K12H4.4 hypothetical protein; K12948 signal peptidase complex
subunit 3 [EC:3.4.-.-]
Length=180
Score = 85.5 bits (210), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 55/166 (33%), Positives = 83/166 (50%), Gaps = 9/166 (5%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFFLVGPIS-----GSVSAREVYGFGYNYAL 57
M N L+RANA+ + + + S+ FL + V R V + +
Sbjct 1 MHNLLSRANALLAFTLWVMAAVTAACFLSTVFLDYTVPTKLTVNDVKVRNVVDYATDEQ- 59
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
D A L+F++K D +F WN KQLFV++VAEY+S + NQVV++DRI+ V+D
Sbjct 60 QADLATLNFNLKVDFSKIFNWNVKQLFVYLVAEYKSKVNEVNQVVLWDRIVERADRVVMD 119
Query 118 LANVPAKYHLRDKGKG-LRGREITLKLQVVYHPIVGRIYTQTVASS 162
V +KY+ D G L + +T L+ Y+ I Y + V SS
Sbjct 120 EIGVKSKYYFLDDGTNLLNHKNVTFVLR--YNVIPNSGYLRLVQSS 163
> xla:432311 spcs3, MGC79052; signal peptidase complex subunit
3 homolog; K12948 signal peptidase complex subunit 3 [EC:3.4.-.-]
Length=180
Score = 82.4 bits (202), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 55/156 (35%), Positives = 80/156 (51%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVS---AREVYGFGYNYAL 57
M L+RAN++F + + L G ++ F + P++ VS V F
Sbjct 1 MNTVLSRANSLFAFSLSVMAALTFGCFITTAFKERIVPVNIHVSRVMLENVEDFTGPRER 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++FDI ADL+ +F WN KQLF+++ AEY + + NQVV++D+II L
Sbjct 61 S-DLGFITFDINADLQPIFDWNVKQLFIYLSAEYSTRSNTLNQVVLWDKIILRGDNPKLS 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L + +KY D G GL+G R ITL L P G
Sbjct 120 LKEMKSKYFFFDDGNGLKGNRNITLTLSWNVVPNAG 155
> sce:YLR066W SPC3; Spc3p; K12948 signal peptidase complex subunit
3 [EC:3.4.-.-]
Length=184
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 41/123 (33%), Positives = 70/123 (56%), Gaps = 2/123 (1%)
Query 43 VSAREVYGFGYNYALSGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVV 102
++ R FG + + + FD+ DL LF WN KQ+FV++ AEY S + T++V
Sbjct 53 INVRTSRYFGSQRGKAKENMKIKFDLNTDLTPLFNWNTKQVFVYLTAEYNSTEKITSEVT 112
Query 103 VFDRIITNESEAVLDLANVPAKYHLRDKGKG-LRGREITLKLQVVYHPIVGRI-YTQTVA 160
+D+II ++ +AV+D+ ++ +KY + D G G+++ KL P VG + Y +TV
Sbjct 113 FWDKIIKSKDDAVIDVNDLRSKYSIWDIEDGKFEGKDLVFKLHWNVQPWVGLLTYGETVG 172
Query 161 SST 163
+ T
Sbjct 173 NYT 175
> mmu:76687 Spcs3, 1810011E08Rik; signal peptidase complex subunit
3 homolog (S. cerevisiae); K12948 signal peptidase complex
subunit 3 [EC:3.4.-.-]
Length=180
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/156 (33%), Positives = 78/156 (50%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVS---AREVYGFGYNYAL 57
M L+RAN++F + + L G ++ F P+ VS + V F
Sbjct 1 MNTVLSRANSLFAFSLSVMAALTFGCFITTAFKDRSVPVRLHVSRIMLKNVEDFTGPRER 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++FDI ADL +F WN KQLF+++ AEY + + NQVV++D+I+ L
Sbjct 61 S-DLGFITFDITADLENIFDWNVKQLFLYLSAEYSTKNNALNQVVLWDKIVLRGDNPKLL 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L ++ KY D G GL+G R +TL L P G
Sbjct 120 LKDMKTKYFFFDDGNGLKGNRNVTLTLSWNVVPNAG 155
> hsa:60559 SPCS3, DKFZp564J1864, FLJ22649, PRO3567, SPC22/23,
SPC3, YLR066W; signal peptidase complex subunit 3 homolog (S.
cerevisiae); K12948 signal peptidase complex subunit 3 [EC:3.4.-.-]
Length=180
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/156 (33%), Positives = 78/156 (50%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVS---AREVYGFGYNYAL 57
M L+RAN++F + + L G ++ F P+ VS + V F
Sbjct 1 MNTVLSRANSLFAFSLSVMAALTFGCFITTAFKDRSVPVRLHVSRIMLKNVEDFTGPRER 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++FDI ADL +F WN KQLF+++ AEY + + NQVV++D+I+ L
Sbjct 61 S-DLGFITFDITADLENIFDWNVKQLFLYLSAEYSTKNNALNQVVLWDKIVLRGDNPKLL 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L ++ KY D G GL+G R +TL L P G
Sbjct 120 LKDMKTKYFFFDDGNGLKGNRNVTLTLSWNVVPNAG 155
> dre:405884 spcs3, MGC85675, zgc:85675; signal peptidase complex
subunit 3 homolog (S. cerevisiae); K12948 signal peptidase
complex subunit 3 [EC:3.4.-.-]
Length=180
Score = 75.9 bits (185), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 51/156 (32%), Positives = 80/156 (51%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVSA---REVYGFGYNYAL 57
M L+RAN++F + + L G ++ F P+ VS + V F
Sbjct 1 MNTVLSRANSLFAFSLSVMAALTFGCFITTAFKDRSVPVDIHVSKVMIKNVDDFTGPRER 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++FD+ A+L+ +F WN K+LF+++ AEY + + NQVV++D+I+ L+
Sbjct 61 S-DLGFVTFDLFANLQPIFDWNVKELFLYLTAEYSTKSNTLNQVVLWDKIVLRGDNTKLN 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L +V +KY D G GLR + ITL L P G
Sbjct 120 LKDVKSKYFFFDDGNGLRANKNITLTLSWNVVPNAG 155
> ath:AT3G05230 signal peptidase subunit family protein
Length=136
Score = 69.7 bits (169), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 67/129 (51%), Gaps = 4/129 (3%)
Query 3 MENYLNRANAVFCSLMVSLG-VLALGNIASSFFLVGPISGSVSAREVYGFGYNYALSGDQ 61
M + RANA+ + +L + A+ + + F P S + + F + D+
Sbjct 1 MHTFGYRANALLTFAVTALAFICAIASFSDKFSNQNP-SAEIQILNINRFK-KQSHGNDE 58
Query 62 AVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLANV 121
L+ DI ADL+ LF WN KQ+FVFV AEYE+P++ NQV ++D II + A + V
Sbjct 59 VSLTLDISADLQSLFTWNTKQVFVFVAAEYETPKNSLNQVSLWDAIIPAKEHAKFRI-QV 117
Query 122 PAKYHLRDK 130
KY D+
Sbjct 118 SNKYRFIDQ 126
> dre:30354 lpl, fb62e04, fc49b03, wu:fb62e04, wu:fc49b03; lipoprotein
lipase (EC:3.1.1.34); K01059 lipoprotein lipase [EC:3.1.1.34]
Length=511
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 12/45 (26%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 84 FVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLANVPAKYHLR 128
FV++ + E+ PT + + II N +E ++D ++ +K+ R
Sbjct 19 FVYISSGLETTIDPTAESITLSDIIGNATEWMMDFTDIESKFSFR 63
> cel:Y24F12A.2 hypothetical protein
Length=338
Score = 30.0 bits (66), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 25/105 (23%), Positives = 47/105 (44%), Gaps = 14/105 (13%)
Query 66 FDIKADLRGLFQWNAKQLFVF-----------VVAEYESPQHPTNQVVVFDRIITNESEA 114
FD D G+FQ LF+ + EY + NQ + F+ + ++++
Sbjct 88 FDDSLDPVGVFQKCEALLFIIDAQAELQEPIATLVEYFCRAYKINQNIKFE-VFVHKADG 146
Query 115 VLDLANVPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTV 159
+ + A V K+++ + K +I + LQV YH + IY ++
Sbjct 147 LTEEARVETKFNIYHQVKETIKDQIDVDLQVTYH--LTSIYDHSI 189
> dre:368620 abcc4, MRP, cb1019, si:busm1-52i16.4, si:dz202l16.5,
si:dz52i16.4; ATP-binding cassette, sub-family C (CFTR/MRP),
member 4; K05673 ATP-binding cassette, subfamily C (CFTR/MRP),
member 4
Length=1327
Score = 28.9 bits (63), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 39/97 (40%), Gaps = 24/97 (24%)
Query 64 LSFDIKADLRGLFQWNAKQ--------LFVFVVAEYE--------------SPQHPTNQV 101
L+ L G+FQW +Q V V EY SP P +
Sbjct 989 LALSYAVTLMGMFQWGVRQSAEVENMMTSVERVVEYTELESEAPWETQKRPSPDWPNRGL 1048
Query 102 VVFDRI-ITNESEAVLDLANVPAKYHLRDKGKGLRGR 137
+ FDR+ + S+ + L N+ A + R+K G+ GR
Sbjct 1049 ITFDRVNFSYSSDGPVVLKNISAMFRPREK-VGIVGR 1084
Lambda K H
0.322 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40