bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2865_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
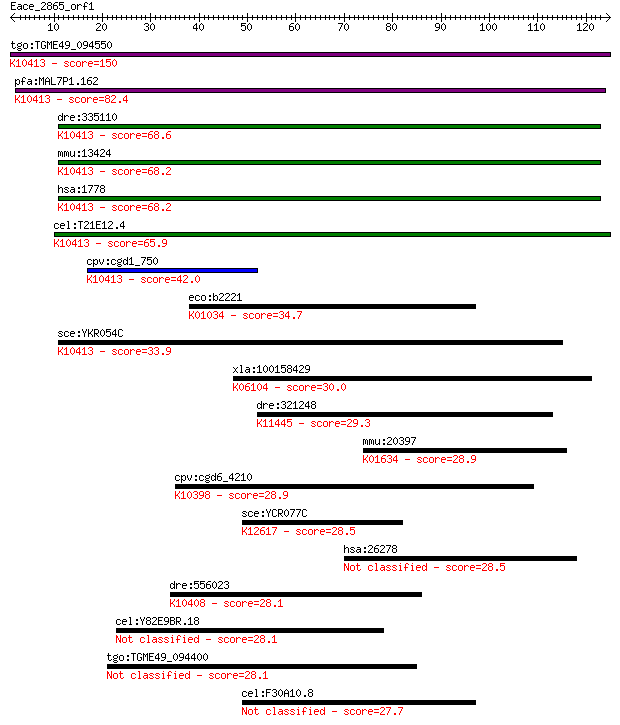
tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein... 150 1e-36
pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein hea... 82.4 3e-16
dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic 1... 68.6 4e-12
mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,... 68.2 6e-12
hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNE... 68.2 6e-12
cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1); ... 65.9 3e-11
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 42.0 5e-04
eco:b2221 atoD, ECK2214, JW2215; acetyl-CoA:acetoacetyl-CoA tr... 34.7 0.075
sce:YKR054C DYN1, DHC1, PAC6; Cytoplasmic heavy chain dynein, ... 33.9 0.13
xla:100158429 amotl1; angiomotin like 1; K06104 angiomotin like 1 30.0
dre:321248 jhdm1da, MGC123306, cb414, kdm7a, sb:cb414, si:dkey... 29.3 2.9
mmu:20397 Sgpl1, AI428538, D10Xrf456, S1PL, Spl; sphingosine p... 28.9 4.5
cpv:cgd6_4210 kinesin-like boursin ; K10398 kinesin family mem... 28.9 4.5
sce:YCR077C PAT1, MRT1; Pat1p; K12617 DNA topoisomerase 2-asso... 28.5 5.8
hsa:26278 SACS, ARSACS, DKFZp686B15167, DNAJC29; spastic ataxi... 28.5 5.8
dre:556023 dynein, axonemal, heavy chain 2-like; K10408 dynein... 28.1 6.6
cel:Y82E9BR.18 hypothetical protein 28.1 7.8
tgo:TGME49_094400 hypothetical protein 28.1 8.1
cel:F30A10.8 stn-1; SynTrophiN family member (stn-1) 27.7 9.8
> tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein
heavy chain 1, cytosolic
Length=4937
Score = 150 bits (378), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 66/124 (53%), Positives = 91/124 (73%), Gaps = 0/124 (0%)
Query 1 SKLEPKWSATIDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVR 60
SK + +W T ++LRASERLLERQR+ FPSDWLW DR+EGE+E F QLLRHQ +VEQ R
Sbjct 1252 SKADVEWEVTFESLRASERLLERQRFAFPSDWLWIDRVEGELETFQQLLRHQALLVEQSR 1311
Query 61 EQLIALVTQFAATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSDYEC 120
E ++ +V ++ A +Q +L+ LYS+WM+ RPV+ D++P HA +L YE QL ++ Y
Sbjct 1312 EPIVDMVKRYNARVQFRLKHLYSEWMIHRPVKSDVSPQHATQVLEGYESQLNQLAEQYAF 1371
Query 121 GKKA 124
G+KA
Sbjct 1372 GEKA 1375
> pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein heavy
chain 1, cytosolic
Length=4985
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 38/122 (31%), Positives = 71/122 (58%), Gaps = 0/122 (0%)
Query 2 KLEPKWSATIDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVRE 61
K E +WS D L+ SE LLE+QRYTFP++WL+ D I G++E Q+ ++Q +++
Sbjct 1262 KEELEWSKKCDLLKQSEVLLEKQRYTFPTNWLYIDNIIGKLETVKQICKYQIKLIKDYLP 1321
Query 62 QLIALVTQFAATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSDYECG 121
+ ++V F +Q +++L+ +W +P + A+ I+ +E++++ I YE
Sbjct 1322 YIQSMVLDFDRKVQNNIKELFEEWNKNKPSHGNANSTKALQIITTFEERIDIINEQYEIS 1381
Query 122 KK 123
+K
Sbjct 1382 EK 1383
> dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4643
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 57/113 (50%), Gaps = 1/113 (0%)
Query 11 IDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVREQLIALVTQF 70
+D R +RLLE+QR+ FP WL+ D IEGE F +++ + ++Q L + Q
Sbjct 1186 VDLFRNGQRLLEKQRFQFPPSWLYIDNIEGEWGAFSDIMKRKDTAIQQQVANLQMKIVQE 1245
Query 71 AATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSDYE-CGK 122
++ + L +DW +PV + P A+ L YE + R+ D E C K
Sbjct 1246 DRAVENRTTDLLADWEKTKPVAGSLRPEEALQSLTIYEGKFGRLKDDREKCAK 1298
> mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,
Dnchc1, Dnec1, Dnecl, Loa, MAP1C, P22, Swl, mKIAA0325; dynein
cytoplasmic 1 heavy chain 1; K10413 dynein heavy chain
1, cytosolic
Length=4644
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 58/113 (51%), Gaps = 1/113 (0%)
Query 11 IDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVREQLIALVTQF 70
++ R +RLLE+QR+ FP WL+ D IEGE F ++R + ++Q L + Q
Sbjct 1185 VELYRNGQRLLEKQRFQFPPSWLYIDNIEGEWGAFNDIMRRKDSAIQQQVANLQMKIVQE 1244
Query 71 AATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSDYE-CGK 122
++ + L +DW +PV ++ P A+ L YE + R+ D E C K
Sbjct 1245 DRAVESRTTDLLTDWEKTKPVTGNLRPEEALQALTIYEGKFGRLKDDREKCAK 1297
> hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNECL,
DYHC, Dnchc1, HL-3, KIAA0325, p22; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4646
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 58/113 (51%), Gaps = 1/113 (0%)
Query 11 IDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVREQLIALVTQF 70
++ R +RLLE+QR+ FP WL+ D IEGE F ++R + ++Q L + Q
Sbjct 1187 VELYRNGQRLLEKQRFQFPPSWLYIDNIEGEWGAFNDIMRRKDSAIQQQVANLQMKIVQE 1246
Query 71 AATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSDYE-CGK 122
++ + L +DW +PV ++ P A+ L YE + R+ D E C K
Sbjct 1247 DRAVESRTTDLLTDWEKTKPVTGNLRPEEALQALTIYEGKFGRLKDDREKCAK 1299
> cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1);
K10413 dynein heavy chain 1, cytosolic
Length=4568
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 68/118 (57%), Gaps = 6/118 (5%)
Query 10 TIDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVREQLIALVTQ 69
+D R+S+RLL +QRY FP+ WL+++ +EGE F ++L + ++ Q++ L T+
Sbjct 1153 AVDLYRSSQRLLNQQRYQFPAQWLYSENVEGEWSAFTEILSLRD---ASIQTQMMNLQTK 1209
Query 70 FA---ATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSDYECGKKA 124
FA ++++ + ++W +PV P A+N++ +E +L ++T + +KA
Sbjct 1210 FAQEDELVEKRTVETLTEWNKSKPVEGAQRPQEALNVITAFEAKLNKLTEERNKMRKA 1267
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 17 SERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRH 51
++ LE QR+TFP+DW+W ++ EGE + Q L H
Sbjct 1335 GQKFLEVQRFTFPNDWIWVEQFEGEYDKSHQRLEH 1369
> eco:b2221 atoD, ECK2214, JW2215; acetyl-CoA:acetoacetyl-CoA
transferase, alpha subunit (EC:2.8.3.8); K01034 acetate CoA-transferase
alpha subunit [EC:2.8.3.8]
Length=220
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 11/67 (16%)
Query 38 IEGEMEVFLQLLRHQGFIVEQVREQLIALV-----TQFAATLQEKLQQLYSD---WMLQR 89
I GEM+V +L QG ++EQ+R L T ++E Q L D W+L+R
Sbjct 89 ISGEMDV---VLVPQGTLIEQIRCGGAGLGGFLTPTGVGTVVEEGKQTLTLDGKTWLLER 145
Query 90 PVRDDIA 96
P+R D+A
Sbjct 146 PLRADLA 152
> sce:YKR054C DYN1, DHC1, PAC6; Cytoplasmic heavy chain dynein,
microtubule motor protein, required for anaphase spindle elongation;
involved in spindle assembly, chromosome movement,
and spindle orientation during cell division, targeted to
microtubule tips by Pac1p; K10413 dynein heavy chain 1, cytosolic
Length=4092
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/104 (18%), Positives = 48/104 (46%), Gaps = 0/104 (0%)
Query 11 IDTLRASERLLERQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVREQLIALVTQF 70
I L + R L + + FPS +++ D+++ + Q L + +++ R + + +
Sbjct 1094 IKLLGSVMRALTKLKVRFPSHFVYIDQLDNDFSSLRQSLSYVEQELQKHRVVIAKSLEEG 1153
Query 71 AATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERI 114
+ Q L W +++P+ + P A+ IL + + + ++
Sbjct 1154 VENINNLSQSLNESWSVRKPISPTLTPPEALKILEFFNESITKL 1197
> xla:100158429 amotl1; angiomotin like 1; K06104 angiomotin like
1
Length=862
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 37/74 (50%), Gaps = 16/74 (21%)
Query 47 QLLRHQGFIVEQVREQLIALVTQFAATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRK 106
QLL + F + + +Q++ +V++ TL++ LQ Y + L+K
Sbjct 330 QLLPPEAFAMVERAQQMVEMVSKENNTLRQHLQTCYDKT----------------DKLQK 373
Query 107 YEQQLERITSDYEC 120
+E ++++I+ YEC
Sbjct 374 FETEIKKISEAYEC 387
> dre:321248 jhdm1da, MGC123306, cb414, kdm7a, sb:cb414, si:dkey-105o6.2,
zgc:123306; jumonji C domain containing histone
demethylase 1 homolog Da (S. cerevisiae); K11445 JmjC domain-containing
histone demethylation protein 1D/E/F
Length=875
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 31/61 (50%), Gaps = 10/61 (16%)
Query 52 QGFIVEQVREQLIALVTQFAATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQL 111
QGF+V V+ AL++ L+ +L Q S+ V D+I PGH + L K + L
Sbjct 392 QGFLVNGVK----ALISSLKMWLRRELTQPNSE------VPDNIRPGHLIKALSKEIRHL 441
Query 112 E 112
E
Sbjct 442 E 442
> mmu:20397 Sgpl1, AI428538, D10Xrf456, S1PL, Spl; sphingosine
phosphate lyase 1 (EC:4.1.2.27); K01634 sphinganine-1-phosphate
aldolase [EC:4.1.2.27]
Length=568
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 5/42 (11%)
Query 74 LQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERIT 115
L E L Q Y ++ P+ DI PG LRK E ++ R+T
Sbjct 156 LTELLVQAYGEFTWSNPLHPDIFPG-----LRKLEAEIVRMT 192
> cpv:cgd6_4210 kinesin-like boursin ; K10398 kinesin family member
11
Length=1184
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 42/88 (47%), Gaps = 14/88 (15%)
Query 35 TDRIEGEMEVFLQ-----LLRHQGFI---VEQVREQLIALVTQFAATL------QEKLQQ 80
T +EG+M+ +L+ L H G I V+ + E+L + T++ + E+L
Sbjct 115 TYTMEGDMKEYLESSNLELTEHAGIIPRAVQLIFERLESQYTEYGVRVSYLEIYNEELSD 174
Query 81 LYSDWMLQRPVRDDIAPGHAMNILRKYE 108
L SD L + DDIA +N+ R E
Sbjct 175 LLSDEKLNLRIYDDIAGKRGLNVDRLEE 202
> sce:YCR077C PAT1, MRT1; Pat1p; K12617 DNA topoisomerase 2-associated
protein PAT1
Length=796
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 24/33 (72%), Gaps = 1/33 (3%)
Query 49 LRHQGFIVEQVREQLIALVTQFAATLQEKLQQL 81
L HQ I+++VR+++ A + + A TLQ+K ++L
Sbjct 717 LNHQRIIIDEVRDEIFATINE-AETLQKKEKEL 748
> hsa:26278 SACS, ARSACS, DKFZp686B15167, DNAJC29; spastic ataxia
of Charlevoix-Saguenay (sacsin)
Length=4579
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 70 FAATLQEKLQQLYSDWMLQRPVRDDIAPGHAMNILRKYEQQLERITSD 117
F L KL + +D+ ++ D+ P + +Y QQLE +T+D
Sbjct 4475 FKCYLSTKLALIAADYAVRGKSDKDVKPTALAQKIEEYSQQLEGLTND 4522
> dre:556023 dynein, axonemal, heavy chain 2-like; K10408 dynein
heavy chain, axonemal
Length=4424
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 34 WTDRIEGEMEVFLQLLRHQGFIVEQVR--EQLIALVTQFAATLQEKLQQLYSDW 85
WT + +E +++LR F+ VR +++ + + TL EK++++++DW
Sbjct 546 WTKALHSRVERSMEVLRRAHFL-PHVRSADEMHVIYVKLCQTLDEKVRRIFTDW 598
> cel:Y82E9BR.18 hypothetical protein
Length=921
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 9/64 (14%)
Query 23 RQRYTFPSDWLWTDRIEGEMEVFLQLLRHQGFIVEQVREQLI---------ALVTQFAAT 73
RQR + + D+ + ++E+ +LL+ QG IVE+V E+ + + +F
Sbjct 740 RQRKEKLAKMKYKDQTDDDLELHKELLKPQGKIVEKVEERPVERPEKSEKPVKIEEFKPK 799
Query 74 LQEK 77
+QEK
Sbjct 800 IQEK 803
> tgo:TGME49_094400 hypothetical protein
Length=1490
Score = 28.1 bits (61), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 15/65 (23%), Positives = 32/65 (49%), Gaps = 1/65 (1%)
Query 21 LERQRYTFPSDWLWTDR-IEGEMEVFLQLLRHQGFIVEQVREQLIALVTQFAATLQEKLQ 79
LE+ + F W ++ ++G + + ++RH F +VRE L++ V + A + +
Sbjct 1006 LEKPQGFFARSWAGVEKTLKGILRLPPYMVRHFVFAAVKVREDLVSEVVREMAAIYNRFS 1065
Query 80 QLYSD 84
+ D
Sbjct 1066 NFFKD 1070
> cel:F30A10.8 stn-1; SynTrophiN family member (stn-1)
Length=440
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 27/55 (49%), Gaps = 7/55 (12%)
Query 49 LRHQGFIVEQVREQLIAL-------VTQFAATLQEKLQQLYSDWMLQRPVRDDIA 96
+RH G+I EQV E I++ +T E + QL ++W R VR +A
Sbjct 228 VRHMGWIAEQVSENGISMWKPKFMTLTNSEILFYEAVPQLKAEWAEPRLVRPLVA 282
Lambda K H
0.321 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40