bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2866_orf1
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
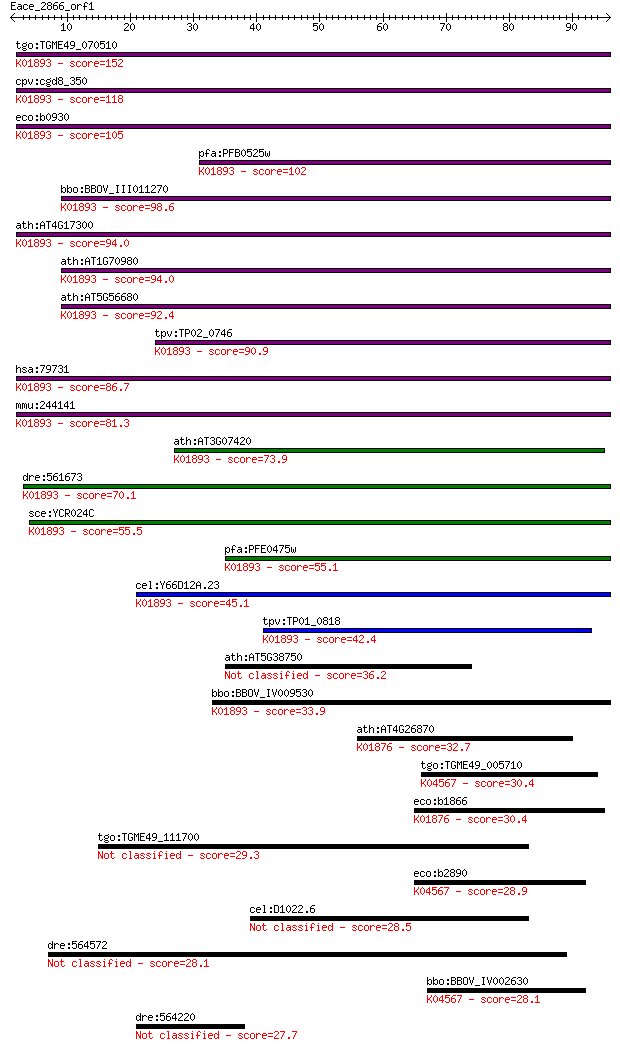
tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.... 152 3e-37
cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase) ;... 118 5e-27
eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA sy... 105 3e-23
pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparagi... 102 3e-22
bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase fami... 98.6 4e-21
ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22); ... 94.0 1e-19
ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligas... 94.0 1e-19
ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligas... 92.4 3e-19
tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 90.9 1e-18
hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase 2... 86.7 2e-17
mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthet... 81.3 7e-16
ath:AT3G07420 NS2; NS2; asparagine-tRNA ligase (EC:6.1.1.22); ... 73.9 1e-13
dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparag... 70.1 1e-12
sce:YCR024C SLM5; Mitochondrial asparaginyl-tRNA synthetase (E... 55.5 4e-08
pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22); K... 55.1 5e-08
cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA s... 45.1 5e-05
tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 42.4 4e-04
ath:AT5G38750 asparaginyl-tRNA synthetase family 36.2 0.024
bbo:BBOV_IV009530 23.m06478; asparaginyl-tRNA synthetase (EC:6... 33.9 0.13
ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--... 32.7 0.33
tgo:TGME49_005710 lysyl-tRNA synthetase, putative (EC:6.1.1.6)... 30.4 1.3
eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase... 30.4 1.6
tgo:TGME49_111700 hypothetical protein 29.3 3.7
eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synth... 28.9 3.7
cel:D1022.6 sro-1; Serpentine Receptor, class O (opsin) family... 28.5 4.9
dre:564572 fc04a02, wu:fc04a02; si:dkey-200l5.5 28.1 6.5
bbo:BBOV_IV002630 21.m02971; lysyl-tRNA synthetase (EC:6.1.1.6... 28.1 6.7
dre:564220 novel protein similar to human membrane-associated ... 27.7 9.3
> tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=676
Score = 152 bits (384), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 70/94 (74%), Positives = 79/94 (84%), Gaps = 0/94 (0%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIG 61
V+SPA GQ +EL V DP +G E+LG DA KYPLAKKEH+REYLREIAHLRPR+ LIG
Sbjct 256 VKSPAKGQAVELAVRDPSKGHRFEILGMTDAAKYPLAKKEHTREYLREIAHLRPRSYLIG 315
Query 62 AVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
AV RVRS LAMATHRFFQDRGFLY+HTPI+T +D
Sbjct 316 AVMRVRSNLAMATHRFFQDRGFLYIHTPIVTASD 349
> cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase)
; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=499
Score = 118 bits (295), Expect = 5e-27, Method: Composition-based stats.
Identities = 52/94 (55%), Positives = 69/94 (73%), Gaps = 2/94 (2%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIG 61
V+SPA GQ +EL + D +++ G DA KYPLAKK HS+E+LRE+AHLRPR+Q
Sbjct 99 VKSPAQGQSVELLLNTGD--DELKICGLCDASKYPLAKKHHSKEFLREVAHLRPRSQFFS 156
Query 62 AVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
+V R+R+ LA+A H +FQ GF+Y+HTPIIT AD
Sbjct 157 SVMRIRNSLAIAIHEYFQKNGFMYIHTPIITAAD 190
> eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=466
Score = 105 bits (263), Expect = 3e-23, Method: Composition-based stats.
Identities = 54/95 (56%), Positives = 64/95 (67%), Gaps = 7/95 (7%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGA-DAGKYPLAKKEHSREYLREIAHLRPRTQLI 60
V SP GQ+ E+Q + VEV G D YP+A K HS EYLRE+AHLRPRT LI
Sbjct 81 VASPGQGQQFEIQAS------KVEVAGWVEDPDTYPMAAKRHSIEYLREVAHLRPRTNLI 134
Query 61 GAVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
GAVARVR LA A HRFF ++GF +V TP+IT +D
Sbjct 135 GAVARVRHTLAQALHRFFNEQGFFWVSTPLITASD 169
> pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=610
Score = 102 bits (254), Expect = 3e-22, Method: Composition-based stats.
Identities = 41/65 (63%), Positives = 54/65 (83%), Gaps = 0/65 (0%)
Query 31 DAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTPI 90
D KYPL+KK H +E+LRE+AHLRPR+ I +V R+R+ L++ATH FFQ RGFLY+HTP+
Sbjct 190 DPQKYPLSKKNHGKEFLREVAHLRPRSYFISSVIRIRNSLSIATHLFFQSRGFLYIHTPL 249
Query 91 ITTAD 95
ITT+D
Sbjct 250 ITTSD 254
> bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase family
protein; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=605
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 50/88 (56%), Positives = 65/88 (73%), Gaps = 2/88 (2%)
Query 9 QKIELQVADPQRGDSVEVLG-GADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVR 67
+K E+ VA P G +EV+G DA KYP+AKK ++E+LRE+AHLRPR+ I +V RVR
Sbjct 179 RKYEVHVA-PLEGHYLEVIGENFDAAKYPIAKKYITQEFLREVAHLRPRSYFISSVMRVR 237
Query 68 SQLAMATHRFFQDRGFLYVHTPIITTAD 95
S LAMATH +FQ G +Y+HTP+ITT D
Sbjct 238 SALAMATHIYFQRLGCIYLHTPLITTTD 265
> ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=567
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 49/94 (52%), Positives = 64/94 (68%), Gaps = 7/94 (7%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIG 61
V S QK+EL+V + + V+G D+ YP+ KK SRE+LR AHLRPRT G
Sbjct 173 VASQGTKQKVELKV------EKIIVVGECDS-SYPIQKKRVSREFLRTKAHLRPRTNTFG 225
Query 62 AVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
AVARVR+ LA ATH+FFQ+ GF++V +PIIT +D
Sbjct 226 AVARVRNTLAYATHKFFQESGFVWVASPIITASD 259
> ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=571
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/87 (52%), Positives = 59/87 (67%), Gaps = 6/87 (6%)
Query 9 QKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRS 68
Q IEL V ++V +G D YPL K + + E+LR++ HLR RT LI AVAR+R+
Sbjct 119 QSIELSV------ETVIAVGTVDPTTYPLPKTKLTPEFLRDVLHLRSRTNLISAVARIRN 172
Query 69 QLAMATHRFFQDRGFLYVHTPIITTAD 95
LA ATH FFQ+ FLY+HTPIITT+D
Sbjct 173 ALAFATHSFFQEHSFLYIHTPIITTSD 199
> ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=572
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 47/87 (54%), Positives = 58/87 (66%), Gaps = 6/87 (6%)
Query 9 QKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRS 68
Q+IEL V V +G DA KYPL K + + E LR++ HLR RT I AVAR+R+
Sbjct 122 QQIELNVV------KVIDVGTVDASKYPLPKTKLTLETLRDVLHLRSRTNSISAVARIRN 175
Query 69 QLAMATHRFFQDRGFLYVHTPIITTAD 95
LA ATH FFQ+ FLY+HTPIITT+D
Sbjct 176 ALAFATHSFFQEHSFLYIHTPIITTSD 202
> tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=710
Score = 90.9 bits (224), Expect = 1e-18, Method: Composition-based stats.
Identities = 42/73 (57%), Positives = 57/73 (78%), Gaps = 1/73 (1%)
Query 24 VEVLGGADA-GKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRG 82
++VLG ++ G YP+AKK+ + E+LRE AHLRPRT LI AV RVRS L++A H FFQ +
Sbjct 253 IQVLGVTNSPGTYPIAKKDLTMEFLRENAHLRPRTYLISAVMRVRSSLSIAIHLFFQSKN 312
Query 83 FLYVHTPIITTAD 95
F Y+++P+ITTAD
Sbjct 313 FHYLNSPVITTAD 325
> hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 39/95 (41%), Positives = 62/95 (65%), Gaps = 7/95 (7%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKE-HSREYLREIAHLRPRTQLI 60
++SP+ Q +EL + + ++V+G DA +P+ KE H EYLR+ H R RT ++
Sbjct 99 IKSPSKRQNVEL------KAEKIKVIGNCDAKDFPIKYKERHPLEYLRQYPHFRCRTNVL 152
Query 61 GAVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
G++ R+RS+ A H FF+D GF+++HTPIIT+ D
Sbjct 153 GSILRIRSEATAAIHSFFKDSGFVHIHTPIITSND 187
> mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthetase
2 (mitochondrial)(putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 38/95 (40%), Positives = 61/95 (64%), Gaps = 7/95 (7%)
Query 2 VESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKE-HSREYLREIAHLRPRTQLI 60
V+S + Q +EL + + +EV+G +A +P+ KE H EYLR+ HLR RT +
Sbjct 99 VKSQSKRQNVEL------KAEKIEVIGDCEAKAFPIKYKERHPLEYLRQYPHLRCRTNAL 152
Query 61 GAVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
G++ RVRS+ A H +F+D GF+++HTP++T+ D
Sbjct 153 GSILRVRSEATAAIHSYFKDNGFVHIHTPVLTSND 187
> ath:AT3G07420 NS2; NS2; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=638
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 33/68 (48%), Positives = 43/68 (63%), Gaps = 0/68 (0%)
Query 27 LGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYV 86
+G D KYPL+KK+ LR+ +H RPRT +G+V RV S L +A+H F Q GF YV
Sbjct 204 VGTVDPEKYPLSKKQLPLHMLRDFSHFRPRTTTVGSVTRVHSALTLASHTFLQYHGFQYV 263
Query 87 HTPIITTA 94
P+ITT
Sbjct 264 QVPVITTT 271
> dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=485
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 35/94 (37%), Positives = 54/94 (57%), Gaps = 7/94 (7%)
Query 3 ESPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKE-HSREYLREIAHLRPRTQLIG 61
+SP Q +ELQ S++V+G + +P KE H EY+R+ HLR RT
Sbjct 107 KSPNKRQNVELQ------AQSIKVVGECNPMDFPFKIKERHPLEYIRQFPHLRCRTNAFS 160
Query 62 AVARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
++ R+RS+ A FF+D G++ +HTP+IT+ D
Sbjct 161 SMLRIRSEATAAIQSFFRDHGYVQIHTPVITSND 194
> sce:YCR024C SLM5; Mitochondrial asparaginyl-tRNA synthetase
(EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=492
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 31/92 (33%), Positives = 50/92 (54%), Gaps = 3/92 (3%)
Query 4 SPADGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAV 63
+P Q ELQ+ +P + S++++G + YPL KK + YLR + L+ RT + A+
Sbjct 97 TPNRKQPFELQIKNPVK--SIKLVGPV-SENYPLQKKYQTLRYLRSLPTLKYRTAYLSAI 153
Query 64 ARVRSQLAMATHRFFQDRGFLYVHTPIITTAD 95
R+RS + +FQ F V PI+T+ D
Sbjct 154 LRLRSFVEFQFMLYFQKNHFTKVSPPILTSND 185
> pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=722
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 41/62 (66%), Gaps = 1/62 (1%)
Query 35 YPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFF-QDRGFLYVHTPIITT 93
Y ++KK H+++YLR HLR RT+L ++ R++S + T F + + + Y++TPI+T+
Sbjct 303 YVISKKYHTKQYLRNYPHLRARTKLYSSLFRLKSDIIFETFNFLKKKKNYTYINTPILTS 362
Query 94 AD 95
D
Sbjct 363 ND 364
> cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=448
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 4/75 (5%)
Query 21 GDSVEVLGGADAGKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQD 80
+ ++V+G + +Y + S + LR+ HLR R A+ R RS + TH FF
Sbjct 80 AEQLKVVGNDENPRY----DDLSPDNLRKKTHLRARNSKFAALLRARSAIFRETHEFFMS 135
Query 81 RGFLYVHTPIITTAD 95
R F+++ TP +T D
Sbjct 136 RDFIHIDTPKLTRND 150
> tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=558
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 41 EHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTPIIT 92
E+S EYLR+ H R R + ++ +RS + H+FF F V TP +T
Sbjct 224 EYSDEYLRKYTHFRFRNKQFRSIIAIRSAIKSYIHQFFTSNKFTEVDTPCLT 275
> ath:AT5G38750 asparaginyl-tRNA synthetase family
Length=135
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 35 YPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMA 73
Y + K + + EYLR+ + R RT +I AV R+R ++ +A
Sbjct 89 YSIPKSKLTLEYLRDHTYFRARTSMIAAVTRIRHKMCLA 127
> bbo:BBOV_IV009530 23.m06478; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=557
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 33/75 (44%), Gaps = 19/75 (25%)
Query 33 GKYPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRF------------FQD 80
G YP+ YLRE + R R ++ A+ R+RS + + Q
Sbjct 191 GIYPMT-------YLREHVNFRSRNAVMQAIFRIRSAVTEGLYHIMKVCHVTGSLYSLQK 243
Query 81 RGFLYVHTPIITTAD 95
R F+ V TP++TT +
Sbjct 244 RSFINVTTPLLTTVN 258
> ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=532
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 56 RTQLIGAVARVRSQLAMATHRFFQDRGFLYVHTP 89
RT A+ R++ Q+ +A + Q +GFL +HTP
Sbjct 219 RTPANQAIFRIQCQVQIAFREYLQSKGFLEIHTP 252
> tgo:TGME49_005710 lysyl-tRNA synthetase, putative (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=658
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 66 VRSQLAMATHRFFQDRGFLYVHTPIITT 93
+RS++ RF +DRGF+ V TP++
Sbjct 305 IRSRIVNYLRRFLEDRGFVEVETPMMNV 332
> eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=590
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 65 RVRSQLAMATHRFFQDRGFLYVHTPIITTA 94
+ R+++ RF D GFL + TP++T A
Sbjct 139 KTRAKITSLVRRFMDDHGFLDIETPMLTKA 168
> tgo:TGME49_111700 hypothetical protein
Length=1280
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 4/72 (5%)
Query 15 VADPQRGDSVEVLGGADAGK----YPLAKKEHSREYLREIAHLRPRTQLIGAVARVRSQL 70
+P+R D G DA + K+E +++ L++ L PR G R R+
Sbjct 634 ATEPRRNDEAGEFNGPDAAANQAPFGFTKREEAQQGLQDSPGLPPRNAEPGNSRRERAAC 693
Query 71 AMATHRFFQDRG 82
++R Q RG
Sbjct 694 RSTSNRLGQARG 705
> eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synthetase,
constitutive (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=505
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 65 RVRSQLAMATHRFFQDRGFLYVHTPII 91
+VRSQ+ +F +RGF+ V TP++
Sbjct 185 KVRSQILSGIRQFMVNRGFMEVETPMM 211
> cel:D1022.6 sro-1; Serpentine Receptor, class O (opsin) family
member (sro-1)
Length=425
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 12/44 (27%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 39 KKEHSREYLREIAHLRPRTQLIGAVARVRSQLAMATHRFFQDRG 82
+++ +YLR+ A L+P L+ + +QL + H F ++G
Sbjct 76 RRQQKGKYLRDNASLQPLVWLLIVDVLMSAQLVVPIHNFLTEKG 119
> dre:564572 fc04a02, wu:fc04a02; si:dkey-200l5.5
Length=402
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 27/101 (26%), Positives = 39/101 (38%), Gaps = 23/101 (22%)
Query 7 DGQKIELQVADPQRGDSVEVLGGADAGKYPLAKKEHSREYL------------------R 48
D I L V D EV+ A YP+ +SR+Y+ R
Sbjct 230 DALVIRLDVVDRDVNTGTEVVHWATHFPYPM----YSRDYVYVRRYQVDLENNLMILVSR 285
Query 49 EIAH-LRPRTQLIGAVARVRSQLAMATHRFFQDRGFLYVHT 88
+ H P TQ V +S++ + HR F + GF Y+ T
Sbjct 286 AVKHPCVPETQEFVRVHSYQSKMVIRPHRSFDENGFDYLLT 326
> bbo:BBOV_IV002630 21.m02971; lysyl-tRNA synthetase (EC:6.1.1.6);
K04567 lysyl-tRNA synthetase, class II [EC:6.1.1.6]
Length=548
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 67 RSQLAMATHRFFQDRGFLYVHTPII 91
RS++ T FF RGF+ V TP +
Sbjct 182 RSRIVSYTRNFFSQRGFMEVETPTL 206
> dre:564220 novel protein similar to human membrane-associated
guanylate kinase-related (MAGI-3) (MAGI-3)
Length=1407
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 10/17 (58%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 21 GDSVEVLGGADAGKYPL 37
GD VE+LGGA+ G++P
Sbjct 29 GDVVEILGGAELGEFPF 45
Lambda K H
0.321 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2065224908
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40