bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2921_orf2
Length=186
Score E
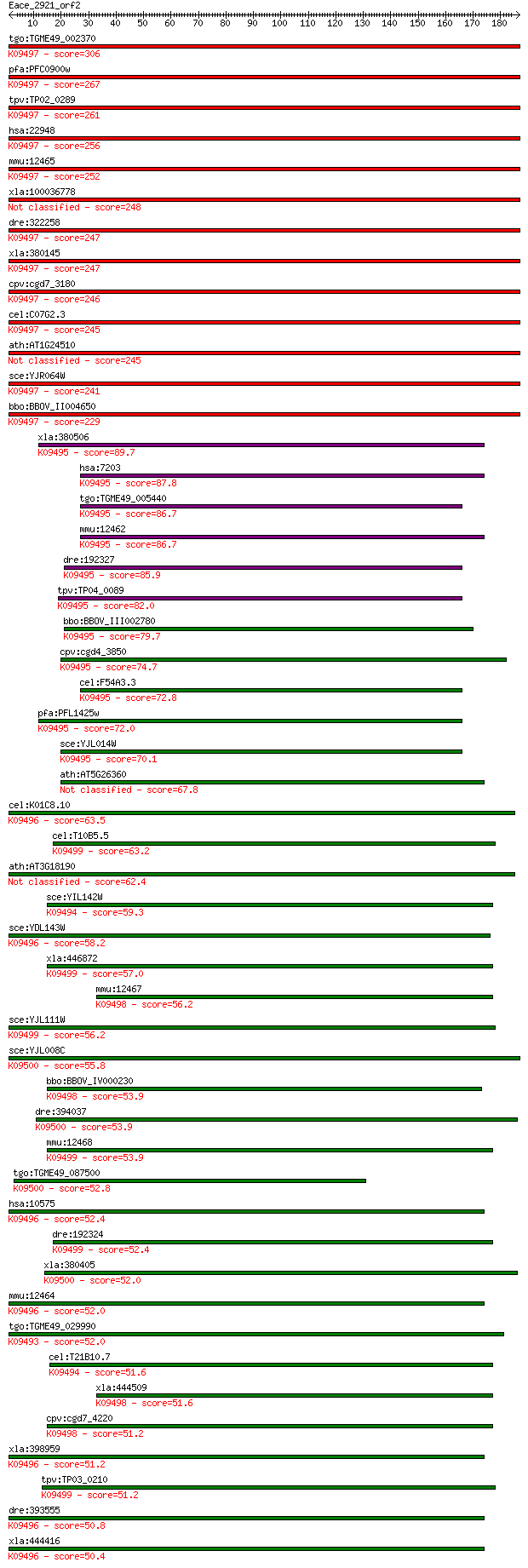
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_002370 TCP-1/cpn60 family chaperonin, putative (EC:... 306 2e-83
pfa:PFC0900w T-complex protein 1 epsilon subunit, putative; K0... 267 1e-71
tpv:TP02_0289 T-complex protein 1 subunit epsilon; K09497 T-co... 261 7e-70
hsa:22948 CCT5, CCT-epsilon, CCTE, KIAA0098, TCP-1-epsilon; ch... 256 3e-68
mmu:12465 Cct5, Ccte, TCPE, mKIAA0098; chaperonin containing T... 252 4e-67
xla:100036778 cct5b; chaperonin containing TCP1, subunit 5 (ep... 248 1e-65
dre:322258 cct5, wu:fb54h08; chaperonin containing TCP1, subun... 247 1e-65
xla:380145 cct5, MGC53061, cct5a; chaperonin containing TCP1, ... 247 2e-65
cpv:cgd7_3180 T complex chaperonin ; K09497 T-complex protein ... 246 3e-65
cel:C07G2.3 cct-5; Chaperonin Containing TCP-1 family member (... 245 6e-65
ath:AT1G24510 T-complex protein 1 epsilon subunit, putative / ... 245 8e-65
sce:YJR064W CCT5, TCP5; Cct5p; K09497 T-complex protein 1 subu... 241 1e-63
bbo:BBOV_II004650 18.m06389; T-complex protein 1 epsilon subun... 229 3e-60
xla:380506 cct3, MGC52575; chaperonin containing TCP-1 complex... 89.7 5e-18
hsa:7203 CCT3, CCT-gamma, CCTG, PIG48, TCP-1-gamma, TRIC5; cha... 87.8 2e-17
tgo:TGME49_005440 TCP-1/cpn60 family chaperonin, putative (EC:... 86.7 4e-17
mmu:12462 Cct3, AL024092, Cctg, Tcp1-rs3, TriC-P5; chaperonin ... 86.7 5e-17
dre:192327 cct3, chunp6930, wu:fb13f04, wu:fb52a02, wu:fj48b06... 85.9 8e-17
tpv:TP04_0089 T-complex protein 1 subunit gamma; K09495 T-comp... 82.0 1e-15
bbo:BBOV_III002780 17.m07264; TCP-1/cpn60 chaperonin family pr... 79.7 5e-15
cpv:cgd4_3850 t-complex protein 1, gamma subunit ; K09495 T-co... 74.7 2e-13
cel:F54A3.3 cct-3; Chaperonin Containing TCP-1 family member (... 72.8 7e-13
pfa:PFL1425w t-complex protein 1, gamma subunit, putative; K09... 72.0 1e-12
sce:YJL014W CCT3, BIN2, TCP3; Cct3p; K09495 T-complex protein ... 70.1 4e-12
ath:AT5G26360 chaperonin, putative 67.8 2e-11
cel:K01C8.10 cct-4; Chaperonin Containing TCP-1 family member ... 63.5 4e-10
cel:T10B5.5 cct-7; Chaperonin Containing TCP-1 family member (... 63.2 5e-10
ath:AT3G18190 chaperonin, putative 62.4 8e-10
sce:YIL142W CCT2, BIN3, TCP2; Cct2p; K09494 T-complex protein ... 59.3 7e-09
sce:YDL143W CCT4, ANC2, TCP4; Cct4p; K09496 T-complex protein ... 58.2 2e-08
xla:446872 cct7, MGC80866; chaperonin containing TCP1, subunit... 57.0 3e-08
mmu:12467 Cct6b, CCTzeta-2, Cctz-2; chaperonin containing Tcp1... 56.2 5e-08
sce:YJL111W CCT7, TCP7; Cct7p; K09499 T-complex protein 1 subu... 56.2 6e-08
sce:YJL008C CCT8; Cct8p; K09500 T-complex protein 1 subunit theta 55.8 8e-08
bbo:BBOV_IV000230 21.m02761; T-complex protein 1 zeta subunit;... 53.9 3e-07
dre:394037 cct8, MGC56059, fa22h09, wu:fa22h09, zgc:56059; cha... 53.9 3e-07
mmu:12468 Cct7, AA408524, AL022769, Ccth, Cctz; chaperonin con... 53.9 3e-07
tgo:TGME49_087500 TCP-1/cpn60 family chaperonin, putative ; K0... 52.8 8e-07
hsa:10575 CCT4, CCT-DELTA, Cctd, MGC126164, MGC126165, SRB; ch... 52.4 8e-07
dre:192324 cct7, chunp6934, fb38h02, fc05g05, wu:fb38h02, wu:f... 52.4 8e-07
xla:380405 cct8, MGC53230; chaperonin containing TCP1, subunit... 52.0 1e-06
mmu:12464 Cct4, 2610204B21Rik, A45, C78323, Cctd; chaperonin c... 52.0 1e-06
tgo:TGME49_029990 TCP-1/cpn60 family chaperonin, putative (EC:... 52.0 1e-06
cel:T21B10.7 cct-2; Chaperonin Containing TCP-1 family member ... 51.6 1e-06
xla:444509 cct6a, MGC80682, MGC81949, cct-zeta, cct6; chaperon... 51.6 1e-06
cpv:cgd7_4220 TCP-1 chaperonin ; K09498 T-complex protein 1 su... 51.2 2e-06
xla:398959 MGC83370; hypothetical protein LOC398959; K09496 T-... 51.2 2e-06
tpv:TP03_0210 T-complex protein 1 subunit eta; K09499 T-comple... 51.2 2e-06
dre:393555 cct4, MGC65789, zgc:65789, zgc:77192; chaperonin co... 50.8 3e-06
xla:444416 cct4, MGC82994; chaperonin containing TCP1, subunit... 50.4 3e-06
> tgo:TGME49_002370 TCP-1/cpn60 family chaperonin, putative (EC:2.7.1.150);
K09497 T-complex protein 1 subunit epsilon
Length=536
Score = 306 bits (785), Expect = 2e-83, Method: Compositional matrix adjust.
Identities = 140/186 (75%), Positives = 165/186 (88%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
VQAVL VAD+ER DVN D+IK+DG+AGGLLEQ+ LV G+VLHKDLSHSQM T++AKIA
Sbjct 186 VQAVLTVADRERNDVNFDLIKVDGKAGGLLEQTMLVNGVVLHKDLSHSQMPSVTKNAKIA 245
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLDI DYKKLQAAEQ YF +MI +VK+AGANFV+CQWGFDDE
Sbjct 246 ILTCPFEPPKPKTKHKLDIKTVEDYKKLQAAEQKYFTDMIQKVKEAGANFVICQWGFDDE 305
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLLL +IPAVRWVGGV++E++A+ATQGRIVPR +EL+E+KLG AG ++E+ SGTMGD
Sbjct 306 ANHLLLHENIPAVRWVGGVELELIALATQGRIVPRWEELTEAKLGHAGVVREISSGTMGD 365
Query 181 KMIVIQ 186
KMIVI+
Sbjct 366 KMIVIE 371
> pfa:PFC0900w T-complex protein 1 epsilon subunit, putative;
K09497 T-complex protein 1 subunit epsilon
Length=535
Score = 267 bits (683), Expect = 1e-71, Method: Compositional matrix adjust.
Identities = 119/186 (63%), Positives = 157/186 (84%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V AVL+VAD +RKDV D+IK++G+ GGLLE+STL++GIVL+K+LSHSQM KE R+AKIA
Sbjct 185 VDAVLSVADMKRKDVRFDLIKIEGKTGGLLEESTLIKGIVLNKELSHSQMIKEVRNAKIA 244
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPK KHKL+ITN Y+ LQA E+ YF +M+ +KKAGANFV+CQWGFDDE
Sbjct 245 ILTCPFEPPKPKIKHKLNITNVDAYRDLQAIEKKYFYDMVASLKKAGANFVICQWGFDDE 304
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
AN+LLL +IPA+RWVGGV++E++AIAT G+I+PR +++ ESKLG+A I+E+ GT+ +
Sbjct 305 ANYLLLKENIPAIRWVGGVEMELIAIATGGKIIPRFEDIDESKLGKADLIREISHGTVNN 364
Query 181 KMIVIQ 186
M+ I+
Sbjct 365 PMVYIE 370
> tpv:TP02_0289 T-complex protein 1 subunit epsilon; K09497 T-complex
protein 1 subunit epsilon
Length=550
Score = 261 bits (668), Expect = 7e-70, Method: Compositional matrix adjust.
Identities = 120/186 (64%), Positives = 158/186 (84%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V+AV+AVAD ERKDVNL++IK++ +AGG LEQ+ L++GIVL+KDLSHSQMRK TRDAKIA
Sbjct 185 VRAVMAVADTERKDVNLELIKIESKAGGRLEQTELIQGIVLNKDLSHSQMRKTTRDAKIA 244
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTK+K++I + Y+KLQ EQ YF +M+D+V+K+GANFV+CQWGFDDE
Sbjct 245 ILTCPFEPPKPKTKNKIEIKDVESYEKLQECEQKYFLDMVDRVEKSGANFVICQWGFDDE 304
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLL+ IPAVRWVGGV++E++AIAT G+IV R ++L+ KLG A I+E+ +GT
Sbjct 305 ANHLLMQRGIPAVRWVGGVEMELIAIATGGQIVARFEDLTPEKLGTAKLIREIATGTDHS 364
Query 181 KMIVIQ 186
+++VI+
Sbjct 365 EVVVIE 370
> hsa:22948 CCT5, CCT-epsilon, CCTE, KIAA0098, TCP-1-epsilon;
chaperonin containing TCP1, subunit 5 (epsilon); K09497 T-complex
protein 1 subunit epsilon
Length=541
Score = 256 bits (654), Expect = 3e-68, Method: Compositional matrix adjust.
Identities = 116/186 (62%), Positives = 147/186 (79%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V AVL VAD ER+DV+ ++IK++G+ GG LE + L++G+++ KD SH QM K+ DAKIA
Sbjct 190 VNAVLTVADMERRDVDFELIKVEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVEDAKIA 249
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLD+T+ DYK LQ E+ F+EMI Q+K+ GAN +CQWGFDDE
Sbjct 250 ILTCPFEPPKPKTKHKLDVTSVEDYKALQKYEKEKFEEMIQQIKETGANLAICQWGFDDE 309
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLLL ++PAVRWVGG +IE++AIAT GRIVPR EL+ KLG AG +QE+ GT D
Sbjct 310 ANHLLLQNNLPAVRWVGGPEIELIAIATGGRIVPRFSELTAEKLGFAGLVQEISFGTTKD 369
Query 181 KMIVIQ 186
KM+VI+
Sbjct 370 KMLVIE 375
> mmu:12465 Cct5, Ccte, TCPE, mKIAA0098; chaperonin containing
Tcp1, subunit 5 (epsilon); K09497 T-complex protein 1 subunit
epsilon
Length=541
Score = 252 bits (644), Expect = 4e-67, Method: Compositional matrix adjust.
Identities = 115/186 (61%), Positives = 145/186 (77%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V AVL VAD ER+DV+ ++IK++G+ GG LE + L++G+++ KD SH QM K+ DAKIA
Sbjct 190 VNAVLTVADMERRDVDFELIKVEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVVDAKIA 249
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLD+ + DYK LQ E+ F+EMI Q+K+ GAN +CQWGFDDE
Sbjct 250 ILTCPFEPPKPKTKHKLDVMSVEDYKALQKYEKEKFEEMIKQIKETGANLAICQWGFDDE 309
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLLL +PAVRWVGG +IE++AIAT GRIVPR EL+ KLG AG +QE+ GT D
Sbjct 310 ANHLLLQNGLPAVRWVGGPEIELIAIATGGRIVPRFSELTSEKLGFAGVVQEISFGTTKD 369
Query 181 KMIVIQ 186
KM+VI+
Sbjct 370 KMLVIE 375
> xla:100036778 cct5b; chaperonin containing TCP1, subunit 5 (epsilon)
b
Length=533
Score = 248 bits (632), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 110/186 (59%), Positives = 145/186 (77%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V ++L VAD +RKDV+ ++IK++G+ GG LE + L++G+++ KD SH QM K +DAKIA
Sbjct 190 VNSILTVADMDRKDVDFELIKVEGKVGGKLEDTKLIKGVIVDKDFSHPQMPKVVKDAKIA 249
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLD+T+ DYK LQ E+ F EM+ Q+K GAN +CQWGFDDE
Sbjct 250 ILTCPFEPPKPKTKHKLDVTSVEDYKALQKYEREKFLEMVKQIKDTGANLAICQWGFDDE 309
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLLL ++PA+RWVGG +IE++AIAT GRIVPR EL+ KLG AG ++E+ GT D
Sbjct 310 ANHLLLQNELPAIRWVGGPEIELIAIATGGRIVPRFSELTPEKLGYAGIVKEISFGTTKD 369
Query 181 KMIVIQ 186
+M+VI+
Sbjct 370 RMLVIE 375
> dre:322258 cct5, wu:fb54h08; chaperonin containing TCP1, subunit
5 (epsilon); K09497 T-complex protein 1 subunit epsilon
Length=541
Score = 247 bits (631), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 112/186 (60%), Positives = 147/186 (79%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V A+L VAD ERKDV+ ++IK++G+ GG LE + L++G+++ K+ SH QM K +D KIA
Sbjct 190 VNAILTVADMERKDVDFELIKVEGKVGGKLEDTQLIKGVIVDKEFSHPQMPKVLKDTKIA 249
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLD+T+ DYK LQ E++ F+EMI QVK+ GAN +CQWGFDDE
Sbjct 250 ILTCPFEPPKPKTKHKLDVTSVEDYKALQKYEKDKFQEMIRQVKENGANLAICQWGFDDE 309
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLLL ++PA+RWVGG +IE++AIAT GRIVPR EL+ KLG AG ++E+ GT D
Sbjct 310 ANHLLLQNELPAIRWVGGPEIELIAIATGGRIVPRFCELTPEKLGTAGLVKEICFGTTKD 369
Query 181 KMIVIQ 186
+M+VI+
Sbjct 370 RMLVIE 375
> xla:380145 cct5, MGC53061, cct5a; chaperonin containing TCP1,
subunit 5 (epsilon); K09497 T-complex protein 1 subunit epsilon
Length=541
Score = 247 bits (630), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 111/186 (59%), Positives = 145/186 (77%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V ++L VAD +RKDV+ ++IK++G+ GG LE + L++G+++ KD SH QM K +DAKIA
Sbjct 190 VNSILTVADMDRKDVDFELIKVEGKVGGKLEDTKLIKGVIVDKDFSHPQMPKVVKDAKIA 249
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLD+T+ DYK LQ E+ F EM+ Q+K GAN +CQWGFDDE
Sbjct 250 ILTCPFEPPKPKTKHKLDVTSVEDYKALQKYEREKFLEMVKQIKDTGANLAICQWGFDDE 309
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLLL ++PAVRWVGG +IE++AIAT GRIVPR EL+ KLG AG ++E+ GT D
Sbjct 310 ANHLLLQNELPAVRWVGGPEIELIAIATGGRIVPRFCELTPEKLGYAGIVKEISFGTTKD 369
Query 181 KMIVIQ 186
+M+VI+
Sbjct 370 RMLVIE 375
> cpv:cgd7_3180 T complex chaperonin ; K09497 T-complex protein
1 subunit epsilon
Length=556
Score = 246 bits (628), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 112/186 (60%), Positives = 146/186 (78%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V AVLAVAD +R+DVN D+I + G+ GG LE+S +V GIVL K++SH QM K+ +AKIA
Sbjct 206 VDAVLAVADLDRRDVNFDLINIQGKPGGRLEESCIVNGIVLDKEMSHPQMNKKIENAKIA 265
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
+LTCPFEPPKPKTKHK+DI +++D+ L EQ YF EMID+V +GAN VVCQWGFDDE
Sbjct 266 LLTCPFEPPKPKTKHKIDIKSSSDFDVLFETEQKYFSEMIDKVVSSGANMVVCQWGFDDE 325
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANH+L +P++RWVGGV+IE+LAIAT IVPR ++L+E+KLG A ++E+ SGT D
Sbjct 326 ANHMLSQRGLPSIRWVGGVEIELLAIATGAHIVPRFEDLAENKLGSAALVKEVTSGTEKD 385
Query 181 KMIVIQ 186
K+I I+
Sbjct 386 KLIFIE 391
> cel:C07G2.3 cct-5; Chaperonin Containing TCP-1 family member
(cct-5); K09497 T-complex protein 1 subunit epsilon
Length=542
Score = 245 bits (626), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 114/186 (61%), Positives = 144/186 (77%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V AVL+VAD E KDVN +MIK++G+ GG LE + LV+GIV+ K +SH QM KE ++AK+A
Sbjct 189 VDAVLSVADIESKDVNFEMIKMEGKVGGRLEDTILVKGIVIDKTMSHPQMPKELKNAKVA 248
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHKLDIT+ D+K L+ E+ F+ MI QVK++GA +CQWGFDDE
Sbjct 249 ILTCPFEPPKPKTKHKLDITSTEDFKALRDYERETFETMIRQVKESGATLAICQWGFDDE 308
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLL D+PAVRWVGG +IE+LAIAT RIVPR ELS+ KLG AG ++E+ G D
Sbjct 309 ANHLLQANDLPAVRWVGGPEIELLAIATNARIVPRFSELSKEKLGTAGLVREITFGAAKD 368
Query 181 KMIVIQ 186
+M+ I+
Sbjct 369 RMLSIE 374
> ath:AT1G24510 T-complex protein 1 epsilon subunit, putative
/ TCP-1-epsilon, putative / chaperonin, putative
Length=535
Score = 245 bits (625), Expect = 8e-65, Method: Compositional matrix adjust.
Identities = 111/186 (59%), Positives = 145/186 (77%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V+AVLAVAD ER+DVNLD+IK++G+ GG LE + L+ GI++ KD+SH QM K+ DA IA
Sbjct 186 VKAVLAVADLERRDVNLDLIKVEGKVGGKLEDTELIYGILIDKDMSHPQMPKQIEDAHIA 245
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
ILTCPFEPPKPKTKHK+DI ++ L+ EQ YF EM+ + K GA V+CQWGFDDE
Sbjct 246 ILTCPFEPPKPKTKHKVDIDTVEKFETLRKQEQQYFDEMVQKCKDVGATLVICQWGFDDE 305
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLL+ ++PAVRWVGGV++E++AIAT GRIVPR QEL+ KLG+AG ++E GT +
Sbjct 306 ANHLLMHRNLPAVRWVGGVELELIAIATGGRIVPRFQELTPEKLGKAGVVREKSFGTTKE 365
Query 181 KMIVIQ 186
+M+ I+
Sbjct 366 RMLYIE 371
> sce:YJR064W CCT5, TCP5; Cct5p; K09497 T-complex protein 1 subunit
epsilon
Length=562
Score = 241 bits (615), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 116/192 (60%), Positives = 145/192 (75%), Gaps = 6/192 (3%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRK-----ETR 55
V+AV+ V DK+RKDV+ D+IK+ GR GG + S L+ G++L KD SH QM K E
Sbjct 205 VEAVINVMDKDRKDVDFDLIKMQGRVGGSISDSKLINGVILDKDFSHPQMPKCVLPKEGS 264
Query 56 DA-KIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQ 114
D K+AILTCPFEPPKPKTKHKLDI++ +Y+KLQ EQ+ FKEMID VKKAGA+ V+CQ
Sbjct 265 DGVKLAILTCPFEPPKPKTKHKLDISSVEEYQKLQTYEQDKFKEMIDDVKKAGADVVICQ 324
Query 115 WGFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELP 174
WGFDDEANHLLL D+PAVRWVGG ++E +AI+T GRIVPR Q+LS+ KLG I E
Sbjct 325 WGFDDEANHLLLQNDLPAVRWVGGQELEHIAISTNGRIVPRFQDLSKDKLGTCSRIYEQE 384
Query 175 SGTMGDKMIVIQ 186
GT D+M++I+
Sbjct 385 FGTTKDRMLIIE 396
> bbo:BBOV_II004650 18.m06389; T-complex protein 1 epsilon subunit;
K09497 T-complex protein 1 subunit epsilon
Length=538
Score = 229 bits (585), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 107/186 (57%), Positives = 143/186 (76%), Gaps = 0/186 (0%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIA 60
V AVL+VAD ERKDVNL++IK++ + GG LE + LVRGIVL K+ SHSQM+K DA+IA
Sbjct 190 VDAVLSVADLERKDVNLELIKIESKTGGRLEDTCLVRGIVLPKEFSHSQMKKTVTDARIA 249
Query 61 ILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDE 120
IL+CPFEPPK KTK +++I + DY+KLQ E+ YF+ M+ +V + AN V+CQWGFDDE
Sbjct 250 ILSCPFEPPKTKTKSRIEIGSVDDYEKLQECEKLYFENMVKRVVDSTANVVICQWGFDDE 309
Query 121 ANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGD 180
ANHLL+ I AVRWVGG ++E++AIAT G+IV R ++L ESKLG AG ++E+ +GT
Sbjct 310 ANHLLMKHGISAVRWVGGAELELVAIATGGKIVGRFEDLDESKLGHAGLVREVATGTEHQ 369
Query 181 KMIVIQ 186
+ I I+
Sbjct 370 QFIYIE 375
> xla:380506 cct3, MGC52575; chaperonin containing TCP-1 complex
gamma chain; K09495 T-complex protein 1 subunit gamma
Length=547
Score = 89.7 bits (221), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 46/163 (28%), Positives = 93/163 (57%), Gaps = 1/163 (0%)
Query 12 RKDVNLD-MIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPK 70
RK++++ K++ GG++E S ++RG++++KD++H +MR+ ++ +I +L C E K
Sbjct 189 RKEIDIKKYAKVEKIPGGIIEDSCVLRGVMVNKDVTHPKMRRLIKNPRIILLDCSLEYKK 248
Query 71 PKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADI 130
+++ +++IT D+ ++ E+ Y +++ + + + + V+ + G D A H L+ A+I
Sbjct 249 GESQTEIEITREEDFARILQMEEEYIQQVCEDIIRLKPDVVITEKGISDLAQHYLVKANI 308
Query 131 PAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
AVR V +A A RI R EL E +G + E+
Sbjct 309 TAVRRVRKTDNNRIARACGARIASRTDELREEDVGTGAGLFEI 351
> hsa:7203 CCT3, CCT-gamma, CCTG, PIG48, TCP-1-gamma, TRIC5; chaperonin
containing TCP1, subunit 3 (gamma); K09495 T-complex
protein 1 subunit gamma
Length=507
Score = 87.8 bits (216), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 42/147 (28%), Positives = 84/147 (57%), Gaps = 0/147 (0%)
Query 27 GGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDITNAADYK 86
GG++E S ++RG++++KD++H +MR+ ++ +I +L E K +++ ++IT D+
Sbjct 168 GGIIEDSCVLRGVMINKDVTHPRMRRYIKNPRIVLLDSSLEYKKGESQTDIEITREEDFT 227
Query 87 KLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGVQIEMLAI 146
++ E+ Y +++ + + + + V+ + G D A H L+ A+I A+R V +A
Sbjct 228 RILQMEEEYIQQLCEDIIQLKPDVVITEKGISDLAQHYLMRANITAIRRVRKTDNNRIAR 287
Query 147 ATQGRIVPRAQELSESKLGRAGCIQEL 173
A RIV R +EL E +G + E+
Sbjct 288 ACGARIVSRPEELREDDVGTGAGLLEI 314
> tgo:TGME49_005440 TCP-1/cpn60 family chaperonin, putative (EC:2.7.1.150);
K09495 T-complex protein 1 subunit gamma
Length=556
Score = 86.7 bits (213), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 43/139 (30%), Positives = 79/139 (56%), Gaps = 0/139 (0%)
Query 27 GGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDITNAADYK 86
GG LE+S ++ G++++KD++H++MR+ + K+ +L CP E K +++ ++IT ++
Sbjct 209 GGDLEESRVLDGVMVNKDVTHAKMRRYIANPKVLLLDCPLEYKKGESQTYVEITKEDEWA 268
Query 87 KLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGVQIEMLAI 146
KL E+ + M D + +G N V+ + G D A H L+ A I +R V +A
Sbjct 269 KLLEQEEKEVRAMCDDIIASGCNLVITEKGVSDLAQHFLVKAGISCIRRVRKTDNNRIAR 328
Query 147 ATQGRIVPRAQELSESKLG 165
T IV R +E+++ +G
Sbjct 329 VTGATIVNRTEEITKEDVG 347
> mmu:12462 Cct3, AL024092, Cctg, Tcp1-rs3, TriC-P5; chaperonin
containing Tcp1, subunit 3 (gamma); K09495 T-complex protein
1 subunit gamma
Length=545
Score = 86.7 bits (213), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 83/147 (56%), Gaps = 0/147 (0%)
Query 27 GGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDITNAADYK 86
GG++E S ++RG++++KD++H +MR+ ++ +I +L E K +++ ++IT D+
Sbjct 206 GGIIEDSCVLRGVMINKDVTHPRMRRYIKNPRIVLLDSSLEYKKGESQTDIEITREEDFT 265
Query 87 KLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGVQIEMLAI 146
++ E+ Y ++ + + + + V+ + G D A H L+ A++ A+R V +A
Sbjct 266 RILQMEEEYIHQLCEDIIQLKPDVVITEKGISDLAQHYLMRANVTAIRRVRKTDNNRIAR 325
Query 147 ATQGRIVPRAQELSESKLGRAGCIQEL 173
A RIV R +EL E +G + E+
Sbjct 326 ACGARIVSRPEELREDDVGTGAGLLEI 352
> dre:192327 cct3, chunp6930, wu:fb13f04, wu:fb52a02, wu:fj48b06;
chaperonin containing TCP1, subunit 3 (gamma); K09495 T-complex
protein 1 subunit gamma
Length=543
Score = 85.9 bits (211), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 39/145 (26%), Positives = 83/145 (57%), Gaps = 0/145 (0%)
Query 21 KLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDIT 80
K++ GG++E S ++RG++++KD++H +MR+ ++ +I +L C E K +++ ++I
Sbjct 199 KVEKVPGGIIEDSCVLRGVMVNKDVTHPRMRRLIKNPRIVLLDCSLEYKKGESQTDIEIA 258
Query 81 NAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGVQ 140
D+ ++ E+ Y +++ + + + + + + G D A H L+ A+I A+R +
Sbjct 259 REEDFARILQMEEEYVQQICEDIIRLKPDLIFTEKGISDLAQHYLMKANITAIRRIRKTD 318
Query 141 IEMLAIATQGRIVPRAQELSESKLG 165
+A A RI R EL+E+ +G
Sbjct 319 NNRIARACGARIASRTDELTENDVG 343
> tpv:TP04_0089 T-complex protein 1 subunit gamma; K09495 T-complex
protein 1 subunit gamma
Length=564
Score = 82.0 bits (201), Expect = 1e-15, Method: Composition-based stats.
Identities = 45/147 (30%), Positives = 79/147 (53%), Gaps = 0/147 (0%)
Query 19 MIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLD 78
+IK++ GG +E S ++ G+V++KD+ H+ M + + +I IL C E K +++ +D
Sbjct 218 LIKVEKIIGGYIEDSKVLDGVVVNKDVVHANMSRRIENPRILILDCTLEYKKGESQTMVD 277
Query 79 ITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGG 138
I + + + KL E+ K+M + + N VV + G D A H L+ A I +R V
Sbjct 278 IYDESVWNKLLLQEETEIKQMCQYIINSNCNLVVTEKGVSDLAQHYLVKAGISCLRRVRK 337
Query 139 VQIEMLAIATQGRIVPRAQELSESKLG 165
++ A IV R +E++ES +G
Sbjct 338 SDTNRISKACGATIVNRPEEITESDIG 364
> bbo:BBOV_III002780 17.m07264; TCP-1/cpn60 chaperonin family
protein; K09495 T-complex protein 1 subunit gamma
Length=549
Score = 79.7 bits (195), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 46/149 (30%), Positives = 75/149 (50%), Gaps = 1/149 (0%)
Query 21 KLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDIT 80
K++ GG+ E S ++ GI+++KD++H M + + ++ IL C E K +++ +DI+
Sbjct 201 KVEKLPGGMPEDSIVLDGILINKDVTHPSMSRRIENPRVLILDCTLEYKKGESQTIVDIS 260
Query 81 NAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGVQ 140
+ A + KL E+ M + + G N V + G D A H L A I +R +
Sbjct 261 DEAGWAKLLEQEETEIFNMCQNIIQTGCNVCVTEKGVSDLAQHFLSKAGISCIRRIRKTD 320
Query 141 IEMLAIATQGRIVPRAQELSESKLGRAGC 169
LA T IV R +EL S +G GC
Sbjct 321 ANRLARVTGATIVNRTEELLPSDVG-TGC 348
> cpv:cgd4_3850 t-complex protein 1, gamma subunit ; K09495 T-complex
protein 1 subunit gamma
Length=559
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 81/162 (50%), Gaps = 2/162 (1%)
Query 20 IKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDI 79
++++ GG +E S ++ G+V++KD+ H +M++ + K+ +L C E K +++ ++I
Sbjct 201 VRIEKIPGGEIEDSYVLDGVVVNKDVVHPRMKRLIINPKVLLLDCTLEYKKGESQTNVEI 260
Query 80 TNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGV 139
T AD++ L E+ + M + G N V + G D A H L+ A I +R V
Sbjct 261 TKEADWEALLRQEEEEVEAMCKDIIATGCNVVFTEKGVSDLAQHFLVKAGISVIRRVRKS 320
Query 140 QIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGDK 181
+A T I R +EL+ + +G C +GD+
Sbjct 321 DNNRIARVTGATIASRTEELTPNDVGT--CCGRFEVKKIGDE 360
> cel:F54A3.3 cct-3; Chaperonin Containing TCP-1 family member
(cct-3); K09495 T-complex protein 1 subunit gamma
Length=543
Score = 72.8 bits (177), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 40/139 (28%), Positives = 70/139 (50%), Gaps = 0/139 (0%)
Query 27 GGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDITNAADYK 86
GG +E S +V+GIV++KD+ H++MR+ + +I +L C E K +++ L+I D
Sbjct 209 GGRIEDSQVVKGIVVNKDILHAKMRRRIENPRIVLLDCNLEYKKGESQTSLEIMREEDIS 268
Query 87 KLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGVQIEMLAI 146
+ E+ ++ D++ K + V + G D A H LL A I +R + LA
Sbjct 269 AILEQEEQAIRKQCDEIIKLKPDLVFTEKGISDLAQHFLLKAGITCLRRLKKTDNNRLAR 328
Query 147 ATQGRIVPRAQELSESKLG 165
R+V +L + +G
Sbjct 329 VCGARVVHDTSDLRDEDVG 347
> pfa:PFL1425w t-complex protein 1, gamma subunit, putative; K09495
T-complex protein 1 subunit gamma
Length=542
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/155 (24%), Positives = 84/155 (54%), Gaps = 1/155 (0%)
Query 12 RKDVNLD-MIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPK 70
RK++++ K++ GG + S +++G++++KD++H +MR+ ++ +I +L C E K
Sbjct 192 RKEIDIKRYAKVEKIPGGDITDSYVLKGVMINKDITHPKMRRYIKNPRILLLDCTLEYKK 251
Query 71 PKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADI 130
+++ ++I + + +L E+ K+M + + + + V+ + G D A H L+ +I
Sbjct 252 AESQTNVEILDEKTWNELLLQEEIEVKKMCEYIIDSKCDIVITEKGVSDLAQHFLVKKNI 311
Query 131 PAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLG 165
+R V + L + IV R +E+ E +G
Sbjct 312 SVIRRVRKTDLNRLERISGATIVNRCEEIVEGDIG 346
> sce:YJL014W CCT3, BIN2, TCP3; Cct3p; K09495 T-complex protein
1 subunit gamma
Length=534
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 74/146 (50%), Gaps = 0/146 (0%)
Query 20 IKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDI 79
++++ GG + S +++G++L+KD+ H +M + + ++ +L CP E K +++ ++I
Sbjct 204 VRVEKIPGGDVLDSRVLKGVLLNKDVVHPKMSRHIENPRVVLLDCPLEYKKGESQTNIEI 263
Query 80 TNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGGV 139
D+ ++ E+ + M +Q+ V+ + G D A H LL +R V
Sbjct 264 EKEEDWNRILQIEEEQVQLMCEQILAVRPTLVITEKGVSDLAQHYLLKGGCSVLRRVKKS 323
Query 140 QIEMLAIATQGRIVPRAQELSESKLG 165
+A T IV R ++L ES +G
Sbjct 324 DNNRIARVTGATIVNRVEDLKESDVG 349
> ath:AT5G26360 chaperonin, putative
Length=555
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/155 (26%), Positives = 78/155 (50%), Gaps = 1/155 (0%)
Query 20 IKLDGRAGGLLEQSTLVRGIVLHKDL-SHSQMRKETRDAKIAILTCPFEPPKPKTKHKLD 78
IK++ GG E S +++G++ +KD+ + +M+++ + +I +L CP E K + + +
Sbjct 198 IKVEKVPGGQFEDSEVLKGVMFNKDVVAPGKMKRKIVNPRIILLDCPLEYKKGENQTNAE 257
Query 79 ITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVRWVGG 138
+ D++ L E+ Y + + Q+ K + V+ + G D A H A + A+R +
Sbjct 258 LVREEDWEVLLKLEEEYIENICVQILKFKPDLVITEKGLSDLACHYFSKAGVSAIRRLRK 317
Query 139 VQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
+A A IV R EL ES +G + E+
Sbjct 318 TDNNRIAKACGAVIVNRPDELQESDIGTGAGLFEV 352
> cel:K01C8.10 cct-4; Chaperonin Containing TCP-1 family member
(cct-4); K09496 T-complex protein 1 subunit delta
Length=540
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 49/190 (25%), Positives = 86/190 (45%), Gaps = 8/190 (4%)
Query 1 VQAVLAVADKER-KDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKI 59
V AV + + E +VNL MIK+ + G +E+S L+ G ++ + AKI
Sbjct 187 VNAVKKIINSENDSNVNLKMIKIIKKMGDTVEESELIEGALIDQKTMGRGAPTRIEKAKI 246
Query 60 AILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGF-- 117
++ PPK ++++ IT+ A + E+ Y E+ Q+K AG N ++ Q
Sbjct 247 GLIQFQISPPKTDMENQVIITDYAQMDRALKEERQYLLEICKQIKAAGCNVLLIQKSILR 306
Query 118 ---DDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELP 174
++ A H L I ++ + IE + R V + LG A ++E+P
Sbjct 307 DAVNELALHFLAKMKIMCIKDIEREDIEFYSRILGCRPVASVDHFNADALGYADLVEEIP 366
Query 175 SGTMGDKMIV 184
+G GD ++
Sbjct 367 TG--GDGKVI 374
> cel:T10B5.5 cct-7; Chaperonin Containing TCP-1 family member
(cct-7); K09499 T-complex protein 1 subunit eta
Length=535
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 40/164 (24%), Positives = 77/164 (46%), Gaps = 3/164 (1%)
Query 17 LDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMR---KETRDAKIAILTCPFEPPKPKT 73
L+MI + GG L +S L++G+ K S++ K+ + K+A+L E K
Sbjct 190 LNMIGIKKVNGGNLHESRLIKGVAFQKAFSYAGFEMQPKKYSNVKVALLNIELELKAEKE 249
Query 74 KHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAV 133
++ +TN +D++ + AE N + + ++ +GAN V+ + D A D D+
Sbjct 250 NAEMRLTNVSDFQAVVDAEWNILYDKLQKIHDSGANVVLSKLPIGDVATQWFADRDMFCA 309
Query 134 RWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGT 177
+ ++ L A G ++ ++ +S LG+ G E G+
Sbjct 310 GRIPQDDLDRLMSACGGSVLTTVSQIDDSVLGKCGKFYEQQVGS 353
> ath:AT3G18190 chaperonin, putative
Length=536
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 52/193 (26%), Positives = 92/193 (47%), Gaps = 14/193 (7%)
Query 1 VQAVLAVADKERKD-VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQ---MRKETRD 56
V AVL+V D E+ + V+L IK+ + GG ++ + V+G+V K +S + R E +
Sbjct 184 VDAVLSVIDPEKPEIVDLRDIKIVKKLGGTVDDTHTVKGLVFDKKVSRAAGGPTRVE--N 241
Query 57 AKIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWG 116
AKIA++ PPK + + +++ ++ E+NY MI ++K G N ++ Q
Sbjct 242 AKIAVIQFQISPPKTDIEQSIVVSDYTQMDRILKEERNYILGMIKKIKATGCNVLLIQKS 301
Query 117 F-----DDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQ 171
D + H L A I ++ V +IE + + + KLG A ++
Sbjct 302 ILRDAVTDLSLHYLAKAKIMVIKDVERDEIEFVTKTLNCLPIANIEHFRAEKLGHADLVE 361
Query 172 ELPSGTMGDKMIV 184
E ++GD I+
Sbjct 362 E---ASLGDGKIL 371
> sce:YIL142W CCT2, BIN3, TCP2; Cct2p; K09494 T-complex protein
1 subunit beta
Length=527
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 51/165 (30%), Positives = 75/165 (45%), Gaps = 6/165 (3%)
Query 15 VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPK-- 72
NL+ I++ GG L S L G +L K ++Q K +AKI I + K K
Sbjct 188 TNLEHIQIIKILGGKLSDSFLDEGFILAKKFGNNQ-PKRIENAKILIANTTLDTDKVKIF 246
Query 73 -TKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIP 131
TK K+D T A +L+ AE+ K I ++ K G N + + D L D I
Sbjct 247 GTKFKVDST--AKLAQLEKAEREKMKNKIAKISKFGINTFINRQLIYDYPEQLFTDLGIN 304
Query 132 AVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
++ +E LA+ T G +V E S+ KLG I+E+ G
Sbjct 305 SIEHADFEGVERLALVTGGEVVSTFDEPSKCKLGECDVIEEIMLG 349
> sce:YDL143W CCT4, ANC2, TCP4; Cct4p; K09496 T-complex protein
1 subunit delta
Length=528
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 49/183 (26%), Positives = 93/183 (50%), Gaps = 10/183 (5%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQ---MRKETRDA 57
V +VL ++D+ K+V+L+ I+L + GG ++ + ++ G+VL + S RKE A
Sbjct 175 VDSVLKISDENSKNVDLNDIRLVKKVGGTIDDTEMIDGVVLTQTAIKSAGGPTRKE--KA 232
Query 58 KIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGF 117
KI ++ PPKP T++ + + + K+ E+ Y + ++KKA N ++ Q
Sbjct 233 KIGLIQFQISPPKPDTENNIIVNDYRQMDKILKEERAYLLNICKKIKKAKCNVLLIQKSI 292
Query 118 -----DDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQE 172
+D A H L +I V+ + +IE L+ + + + +E +LG A ++E
Sbjct 293 LRDAVNDLALHFLSKLNIMVVKDIEREEIEFLSKGLGCKPIADIELFTEDRLGSADLVEE 352
Query 173 LPS 175
+ S
Sbjct 353 IDS 355
> xla:446872 cct7, MGC80866; chaperonin containing TCP1, subunit
7 (eta); K09499 T-complex protein 1 subunit eta
Length=542
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/165 (26%), Positives = 69/165 (41%), Gaps = 3/165 (1%)
Query 15 VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMR---KETRDAKIAILTCPFEPPKP 71
+ L MI + GG LE S LV G+ K S++ K+ KIA+L E
Sbjct 190 LQLKMIGIKKVQGGALEDSHLVAGVAFKKTFSYAGFEMQPKKYESPKIALLNVELELKAE 249
Query 72 KTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIP 131
K ++ + N DY+ + AE N + ++++ K+GA V+ + D A D D+
Sbjct 250 KDNAEVRVNNIEDYQAIVDAEWNILYDKLEKIYKSGAKVVLSKLPIGDVATQYFADRDLF 309
Query 132 AVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
V ++ +A G I L+ LG +E G
Sbjct 310 CAGRVPEEDLKRTMMACGGSIQTSVNALTHDVLGGCALFEETQVG 354
> mmu:12467 Cct6b, CCTzeta-2, Cctz-2; chaperonin containing Tcp1,
subunit 6b (zeta); K09498 T-complex protein 1 subunit zeta
Length=531
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 43/153 (28%), Positives = 68/153 (44%), Gaps = 9/153 (5%)
Query 33 STLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAE 92
+ L+RG+VL H +MRK+ RDA I E K + + +KL AE
Sbjct 204 TQLIRGLVLDHGARHPRMRKQVRDAYILTCNVSLEYEKTEVSSGFFYKTVEEKEKLVKAE 263
Query 93 QNYFKEMIDQV--------KKAGANFVVC-QWGFDDEANHLLLDADIPAVRWVGGVQIEM 143
+ + ++ + ++ ++ FVV Q G D + +L +I A+R +E
Sbjct 264 RKFIEDRVQKIIDLKQKVCAESNKGFVVINQKGIDPVSLEMLAKHNIVALRRAKRRNLER 323
Query 144 LAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
L +A G V + LSE LG AG + E G
Sbjct 324 LTLACGGLAVNSFEGLSEECLGHAGLVFEYALG 356
> sce:YJL111W CCT7, TCP7; Cct7p; K09499 T-complex protein 1 subunit
eta
Length=550
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 44/180 (24%), Positives = 78/180 (43%), Gaps = 6/180 (3%)
Query 1 VQAVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHS---QMRKETRDA 57
V AVL++ +R D++ +I + GG +E+S + G+ K S++ Q K+ +
Sbjct 183 VDAVLSL---DRNDLDDKLIGIKKIPGGAMEESLFINGVAFKKTFSYAGFEQQPKKFNNP 239
Query 58 KIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGF 117
KI L E K ++ + + DY+ + AE E + QV++ GAN V+ +
Sbjct 240 KILSLNVELELKAEKDNAEVRVEHVEDYQAIVDAEWQLIFEKLRQVEETGANIVLSKLPI 299
Query 118 DDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGT 177
D A D +I V + + A G I ++ LG +E+ G+
Sbjct 300 GDLATQFFADRNIFCAGRVSADDMNRVIQAVGGSIQSTTSDIKPEHLGTCALFEEMQIGS 359
> sce:YJL008C CCT8; Cct8p; K09500 T-complex protein 1 subunit
theta
Length=568
Score = 55.8 bits (133), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 45/189 (23%), Positives = 84/189 (44%), Gaps = 5/189 (2%)
Query 1 VQAVLAVADK--ERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDL-SHSQMRKETRDA 57
V VL VA + E N+D I++ GG L ST+++G+V +++ H + E +
Sbjct 186 VSHVLPVAQQAGEIPYFNVDSIRVVKIMGGSLSNSTVIKGMVFNREPEGHVKSLSEDKKH 245
Query 58 KIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGF 117
K+A+ TCP + +TK + + NA + E+ M+ ++ G +V G
Sbjct 246 KVAVFTCPLDIANTETKGTVLLHNAQEMLDFSKGEEKQIDAMMKEIADMGVECIVAGAGV 305
Query 118 DDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGT 177
+ A H L I ++ ++ L +PR + +LG ++ + G
Sbjct 306 GELALHYLNRYGILVLKVPSKFELRRLCRVCGATPLPRLGAPTPEELGLVETVKTMEIG- 364
Query 178 MGDKMIVIQ 186
GD++ V +
Sbjct 365 -GDRVTVFK 372
> bbo:BBOV_IV000230 21.m02761; T-complex protein 1 zeta subunit;
K09498 T-complex protein 1 subunit zeta
Length=538
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 47/168 (27%), Positives = 76/168 (45%), Gaps = 10/168 (5%)
Query 15 VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTK 74
++L MI++ L ++ LVRG+VL H M K+ ++A I L C E + +
Sbjct 190 LDLFMIEVIHMRHRLASETQLVRGMVLDHGARHPDMPKKIKNAYILTLNCSLEYERSEVH 249
Query 75 HKLDITNAADYKKLQAAEQNYFKE----MIDQVKKA-----GANFVVC-QWGFDDEANHL 124
++A ++L ++E+ + E +ID +K G F V Q G D A +
Sbjct 250 SGFSYSSAEQRERLVSSERKFTDEKVQKIIDLKRKVCTPENGRTFCVLNQKGIDPPALDM 309
Query 125 LLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQE 172
L I A+R V +E L + G ++LSE LG A + E
Sbjct 310 LARDGIVALRRVKRRNMERLTLCCGGNACNSVEDLSEDDLGFAEVVYE 357
> dre:394037 cct8, MGC56059, fa22h09, wu:fa22h09, zgc:56059; chaperonin
containing TCP1, subunit 8 (theta); K09500 T-complex
protein 1 subunit theta
Length=546
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 41/175 (23%), Positives = 73/175 (41%), Gaps = 5/175 (2%)
Query 11 ERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPK 70
E N+D +++ G L ST++ G+V K+ +DAKIA+ +CPF+
Sbjct 193 ESGSFNVDNVRVCKILGCGLNSSTMLHGMVFKKEAEGDV--TSVKDAKIAVFSCPFDCTV 250
Query 71 PKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADI 130
+TK + I NA + E++ + + + +GA+ VV D A H +
Sbjct 251 TETKGTVLIKNADELMNFSKGEEDLMEAQVKAIADSGASVVVTGGKVADMALHYANKYKL 310
Query 131 PAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGDKMIVI 185
VR + L +PR + ++GR + +GD +V+
Sbjct 311 MVVRLNSKWDLRRLCKTVGATALPRLTSPTPEEMGRCDSVY---LSEVGDTQVVV 362
> mmu:12468 Cct7, AA408524, AL022769, Ccth, Cctz; chaperonin containing
Tcp1, subunit 7 (eta); K09499 T-complex protein 1
subunit eta
Length=544
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 40/165 (24%), Positives = 70/165 (42%), Gaps = 3/165 (1%)
Query 15 VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMR---KETRDAKIAILTCPFEPPKP 71
+ L MI + GG LE+S LV G+ K S++ K+ ++ KIA+L E
Sbjct 190 LQLKMIGIKKVQGGALEESQLVAGVAFKKTFSYAGFEMQPKKYKNPKIALLNVELELKAE 249
Query 72 KTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIP 131
K ++ + DY+ + AE N + ++++ ++GA ++ + D A D D+
Sbjct 250 KDNAEIRVHTVEDYQAIVDAEWNILYDKLEKIHQSGAKVILSKLPIGDVATQYFADRDMF 309
Query 132 AVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
V ++ +A G I L LG +E G
Sbjct 310 CAGRVPEEDLKRTMMACGGSIQTSVNALVPDVLGHCQVFEETQIG 354
> tgo:TGME49_087500 TCP-1/cpn60 family chaperonin, putative ;
K09500 T-complex protein 1 subunit theta
Length=556
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 61/129 (47%), Gaps = 3/129 (2%)
Query 3 AVLAVADKERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAIL 62
A + + KD +++ I++ GG L QS +++G+V+ + S S K+ + K+ +L
Sbjct 184 AAMVMPPNNPKDFDVENIRVAKLTGGRLSQSVVLKGMVVTRPPSGSVESKQ--NCKVMVL 241
Query 63 TCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWG-FDDEA 121
C E + K + + NA + K E+ +E+I +K AG ++ G D A
Sbjct 242 GCGLECSTTEAKGTVLVHNAEELKNFTKGEERQMEEIIKGIKDAGVEVIIVHGGAISDVA 301
Query 122 NHLLLDADI 130
H DI
Sbjct 302 QHFCNKYDI 310
> hsa:10575 CCT4, CCT-DELTA, Cctd, MGC126164, MGC126165, SRB;
chaperonin containing TCP1, subunit 4 (delta); K09496 T-complex
protein 1 subunit delta
Length=539
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 44/179 (24%), Positives = 81/179 (45%), Gaps = 7/179 (3%)
Query 1 VQAVLAVAD-KERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKI 59
V AV+ V D V+L IK+ + GG ++ LV G+VL + +S+S + + AKI
Sbjct 188 VNAVMKVIDPATATSVDLRDIKIVKKLGGTIDDCELVEGLVLTQKVSNSGITR-VEKAKI 246
Query 60 AILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQW---- 115
++ PK +++ +++ A ++ E+ Y ++ Q+KK G N ++ Q
Sbjct 247 GLIQFCLSAPKTDMDNQIVVSDYAQMDRVLREERAYILNLVKQIKKTGCNVLLIQKSILR 306
Query 116 -GFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
D A H L I ++ + IE + + V + + LG A +E+
Sbjct 307 DALSDLALHFLNKMKIMVIKDIEREDIEFICKTIGTKPVAHIDQFTADMLGSAELAEEV 365
> dre:192324 cct7, chunp6934, fb38h02, fc05g05, wu:fb38h02, wu:fc05g05;
chaperonin containing TCP1, subunit 7 (eta); K09499
T-complex protein 1 subunit eta
Length=547
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 40/163 (24%), Positives = 71/163 (43%), Gaps = 3/163 (1%)
Query 17 LDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMR---KETRDAKIAILTCPFEPPKPKT 73
L MI + GG LE+S LV G+ K S++ K + KIA+L E K
Sbjct 192 LKMIGVKKVQGGALEESQLVAGVAFKKTFSYAGFEMQPKRYMNPKIALLNIELELKAEKD 251
Query 74 KHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAV 133
++ + + DY+ + AE N + ++++ K+GA V+ + D A ++
Sbjct 252 NAEVRVNSMEDYQAIVDAEWNILYDKLEKIHKSGAKVVLSKLPIGDVATQYFAYRNLFCA 311
Query 134 RWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
V ++ +A G I L++ LG+ +E+ G
Sbjct 312 ARVVEENLKRTMMACGGSIQTSVGSLTDDVLGQCELFEEVQVG 354
> xla:380405 cct8, MGC53230; chaperonin containing TCP1, subunit
8 (theta); K09500 T-complex protein 1 subunit theta
Length=546
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/172 (22%), Positives = 74/172 (43%), Gaps = 5/172 (2%)
Query 14 DVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKT 73
+ N+D I++ G + S+++ G+V K++ +DAKIA+ +CPF+ +T
Sbjct 196 NFNVDNIRVCKILGSGICSSSVLHGMVFKKEVEGD--ITSVKDAKIAVYSCPFDGTITET 253
Query 74 KHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAV 133
K + I +A + E+N +E + + AGA +V D A H ++ V
Sbjct 254 KGTVLINSAQELMNFSKGEENLMEEQVKAIADAGATVIVTGGKVADMALHYANKYNLMVV 313
Query 134 RWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGTMGDKMIVI 185
R + L +PR + ++G + +GD +V+
Sbjct 314 RLNSKWDLRRLCKTVCATALPRMTPPTAEEIGHCDSVY---LSEVGDTQVVV 362
> mmu:12464 Cct4, 2610204B21Rik, A45, C78323, Cctd; chaperonin
containing Tcp1, subunit 4 (delta); K09496 T-complex protein
1 subunit delta
Length=539
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/179 (25%), Positives = 81/179 (45%), Gaps = 7/179 (3%)
Query 1 VQAVLAVAD-KERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKI 59
V AV+ V D V+L IK+ + GG ++ LV G+VL + +++S + + AKI
Sbjct 188 VNAVMKVIDPATATSVDLRDIKIVKKLGGTIDDCELVEGLVLTQKVANSGITR-VEKAKI 246
Query 60 AILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQW---- 115
++ PK +++ +++ A ++ E+ Y ++ Q+KK G N ++ Q
Sbjct 247 GLIQFCLSAPKTDMDNQIVVSDYAQMDRVLREERAYILNLVKQIKKTGCNVLLIQKSILR 306
Query 116 -GFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
D A H L I V+ V IE + + V + + LG A +E+
Sbjct 307 DALSDLALHFLNKMKIMVVKDVEREDIEFICKTIGTKPVAHIDQFTADMLGSAELAEEV 365
> tgo:TGME49_029990 TCP-1/cpn60 family chaperonin, putative (EC:2.7.1.150);
K09493 T-complex protein 1 subunit alpha
Length=548
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 47/189 (24%), Positives = 92/189 (48%), Gaps = 12/189 (6%)
Query 1 VQAVLAV-ADKERKDV-----NLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKET 54
V+A+L+V ER DV ++++IK G++ + +S+LV G L + M +
Sbjct 171 VRAILSVKMITERGDVKYPVSSINIIKTHGKS---MRESSLVEGYALKAGRAAQGMPQCV 227
Query 55 RDAKIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQ 114
++AK+A+L + + ++ + N + +K++ E++ I ++ +GAN ++
Sbjct 228 KNAKVALLDFNLRQHRMQLGVQIQVDNPEELEKIRQKEKDITAAKIQKILASGANVILTT 287
Query 115 WGFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCI---Q 171
G DD A ++A AVR V + +A T+G +V L + A C+ +
Sbjct 288 QGIDDMAMKYFVEAGAIAVRRVDRKDLRRIAKITEGTVVLTMATLDGDEKFDASCLGTCE 347
Query 172 ELPSGTMGD 180
E+ +GD
Sbjct 348 EVYEERIGD 356
> cel:T21B10.7 cct-2; Chaperonin Containing TCP-1 family member
(cct-2); K09494 T-complex protein 1 subunit beta
Length=529
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 71/162 (43%), Gaps = 2/162 (1%)
Query 16 NLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKT-K 74
NLD I++ + GG + +S L G +L K Q R+ AKI I P + K K
Sbjct 189 NLDAIQIIKKLGGSMNESYLDEGFLLEKLPGMFQPRR-VEKAKILIANTPMDTDKVKVFG 247
Query 75 HKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDADIPAVR 134
++ + A +L+AAE+ KE +D++ N + + + L DA + A+
Sbjct 248 SRVRVDGVAKVAELEAAEKLKMKEKVDKILAHNCNVFINRQLIYNYPEQLFADAKVMAIE 307
Query 135 WVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
IE LA+ G IV ++ G I+E+ G
Sbjct 308 HADFEGIERLALVLGGEIVSTFDSPQTAQFGSCDLIEEIMIG 349
> xla:444509 cct6a, MGC80682, MGC81949, cct-zeta, cct6; chaperonin
containing TCP1, subunit 6A (zeta 1); K09498 T-complex
protein 1 subunit zeta
Length=531
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 65/153 (42%), Gaps = 9/153 (5%)
Query 33 STLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTKHKLDITNAADYKKLQAAE 92
+TL+RG+VL H M+K DA I E K + +A + +KL AE
Sbjct 204 TTLIRGLVLDHGARHPDMKKRVEDAFILTCNVSLEYEKTEVNSGFFYKSADEREKLVKAE 263
Query 93 QNYFKEMIDQV--------KKAGANFVVC-QWGFDDEANHLLLDADIPAVRWVGGVQIEM 143
+ + +E ++++ G F+V Q G D + L I A+R +E
Sbjct 264 RTFIEERVNKIIALKHKVCGDTGKGFIVLNQKGIDPFSLDALAKEGIVALRRAKRRNMER 323
Query 144 LAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
L +A G + +L+ LG AG + E G
Sbjct 324 LTLACGGSAMNSVDDLTPECLGHAGLVYEYTLG 356
> cpv:cgd7_4220 TCP-1 chaperonin ; K09498 T-complex protein 1
subunit zeta
Length=532
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/172 (25%), Positives = 74/172 (43%), Gaps = 10/172 (5%)
Query 15 VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKIAILTCPFEPPKPKTK 74
V+L MI++ GL ++ LVRG+V+ H M KE ++ I L E +
Sbjct 189 VDLHMIEILPMKHGLTSETRLVRGMVMDHGARHPDMPKELKNCFILTLNVSLEYENSEVT 248
Query 75 HKLDITNAADYKKLQAAEQNYFKEMIDQV---------KKAGANFVVC-QWGFDDEANHL 124
++A ++L AE+ + E + + + ++FVV Q G D + +
Sbjct 249 SSFKYSSAEQRERLVEAERAFTDEKVKSIIDLKRRVCEQNPNSSFVVLNQKGIDPPSLSM 308
Query 125 LLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSG 176
I A+R V +E L +A G + ++LS LG A + E G
Sbjct 309 FAQEGILALRRVKRRNMERLTLACGGNAMNSVEDLSIEDLGWANHVYERSIG 360
> xla:398959 MGC83370; hypothetical protein LOC398959; K09496
T-complex protein 1 subunit delta
Length=541
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/179 (24%), Positives = 83/179 (46%), Gaps = 7/179 (3%)
Query 1 VQAVLAVADKERKD-VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKI 59
V AV+ V D + + V+L IK+ + GG ++ LV G+VL + ++++ + + AKI
Sbjct 190 VDAVMKVIDPQTANSVDLRDIKIVKKLGGTIDDCELVDGLVLTQKVANTGVTR-VEKAKI 248
Query 60 AILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQW---- 115
++ PK +++ +++ A ++ E+ Y ++ Q+KKAG N ++ Q
Sbjct 249 GLIQFCLSAPKTDMDNQIVVSDYAQMDRVLREERTYILNLVKQIKKAGCNVLLIQKSILR 308
Query 116 -GFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
D A H L I V+ + IE + + V + + L A +E+
Sbjct 309 DALSDLALHFLNKMKIMVVKEIEREDIEFICKTIGTKPVAHIDQFTADMLASAELAEEV 367
> tpv:TP03_0210 T-complex protein 1 subunit eta; K09499 T-complex
protein 1 subunit eta
Length=579
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 46/168 (27%), Positives = 73/168 (43%), Gaps = 3/168 (1%)
Query 13 KDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHS---QMRKETRDAKIAILTCPFEPP 69
+D++ DMI + GG E S LV+G+ K S++ Q K+ + KI +L E
Sbjct 190 EDLDEDMIGVKKVTGGSCEDSLLVKGVAFKKTFSYAGAEQQPKKFVNPKILLLNLELELK 249
Query 70 KPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQWGFDDEANHLLLDAD 129
K ++ I N +Y+K+ AE E ++ K GAN V+ + D A D +
Sbjct 250 SEKENAEIVINNPQEYQKIIDAEYRIIFEKLENAVKLGANVVLSKLPIGDLATQYFADKN 309
Query 130 IPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQELPSGT 177
+ V + + AT I LS LG G +E+ G+
Sbjct 310 VFCAGRVDENDLIRTSKATGASIQTTLNNLSVDVLGTCGVFEEVQIGS 357
> dre:393555 cct4, MGC65789, zgc:65789, zgc:77192; chaperonin
containing TCP1, subunit 4 (delta); K09496 T-complex protein
1 subunit delta
Length=533
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/179 (23%), Positives = 82/179 (45%), Gaps = 7/179 (3%)
Query 1 VQAVLAVAD-KERKDVNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKI 59
V AV+ V D V+L I + + GG ++ LV G+VL + ++ + + + AKI
Sbjct 182 VDAVMKVIDPATATSVDLRDINVVKKLGGTIDDCELVDGMVLTQRVASTGVSR-VEKAKI 240
Query 60 AILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQW---- 115
++ PPK +++ +++ A ++ E+ Y ++ QVKKAG ++ Q
Sbjct 241 GLIQFCLSPPKTDMDNQIVVSDYAQMDRVLREERAYILNLVKQVKKAGCTVLLIQKSILR 300
Query 116 -GFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
D A H L I ++ + +IE + + + + + LG A ++E+
Sbjct 301 DALSDLALHFLNKMKIMVIKDIEREEIEFICKTIGTKPIAHIDQFTPEMLGSAELVEEV 359
> xla:444416 cct4, MGC82994; chaperonin containing TCP1, subunit
4 (delta); K09496 T-complex protein 1 subunit delta
Length=539
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/179 (23%), Positives = 82/179 (45%), Gaps = 7/179 (3%)
Query 1 VQAVLAVADKERKD-VNLDMIKLDGRAGGLLEQSTLVRGIVLHKDLSHSQMRKETRDAKI 59
V AV+ V D + + V+L IK+ + GG ++ LV G+VL + +S++ + + AKI
Sbjct 188 VDAVMKVIDPQTANSVDLRDIKIVKKFGGTIDDCELVDGLVLTQKVSNTGVTR-VEKAKI 246
Query 60 AILTCPFEPPKPKTKHKLDITNAADYKKLQAAEQNYFKEMIDQVKKAGANFVVCQW---- 115
++ PK +++ +++ A ++ E+ Y ++ Q+KK G N ++ Q
Sbjct 247 GLIQFCLSAPKTDMDNQIVVSDYAQMDRVLREERTYILNLVKQIKKVGCNVLLIQKSILR 306
Query 116 -GFDDEANHLLLDADIPAVRWVGGVQIEMLAIATQGRIVPRAQELSESKLGRAGCIQEL 173
D A H L I ++ + IE + + V + + L A +E+
Sbjct 307 DALSDLALHFLNKMKIMVIKDIEREDIEFICKTIGAKPVAHIDQFTADMLASAELAEEV 365
Lambda K H
0.319 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40