bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2928_orf1
Length=89
Score E
Sequences producing significant alignments: (Bits) Value
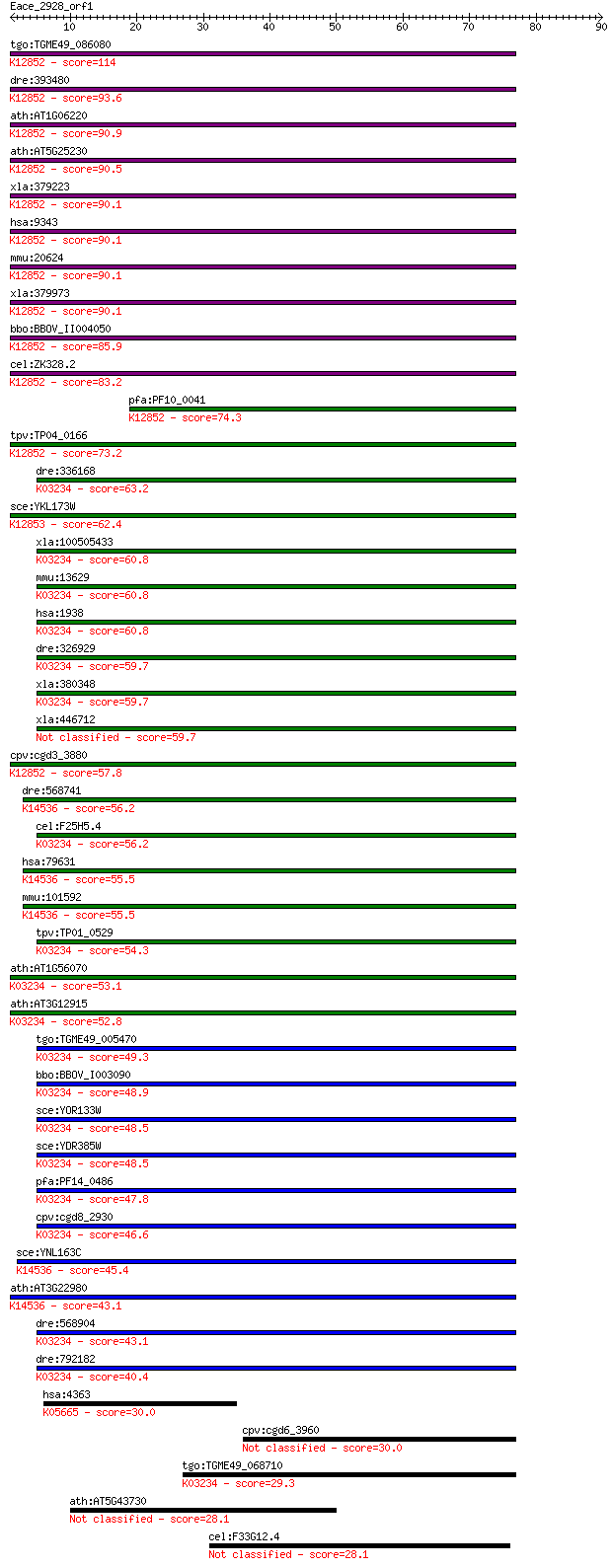
tgo:TGME49_086080 U5 small nuclear ribonucleoprotein, putative... 114 5e-26
dre:393480 eftud2, MGC66214, wu:fi20f05, zgc:66214; elongation... 93.6 1e-19
ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5); GT... 90.9 8e-19
ath:AT5G25230 elongation factor Tu family protein; K12852 116 ... 90.5 1e-18
xla:379223 eftud2, MGC53479, snrp116, snu114; elongation facto... 90.1 1e-18
hsa:9343 EFTUD2, DKFZp686E24196, FLJ44695, KIAA0031, Snrp116, ... 90.1 1e-18
mmu:20624 Eftud2, 116kDa, Snrp116, U5-116kD; elongation factor... 90.1 1e-18
xla:379973 snrp116-pending, MGC52678; U5 snRNP-specific protei... 90.1 1e-18
bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear prot... 85.9 3e-17
cel:ZK328.2 eft-1; Elongation FacTor family member (eft-1); K1... 83.2 2e-16
pfa:PF10_0041 U5 small nuclear ribonuclear protein, putative; ... 74.3 9e-14
tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116 k... 73.2 2e-13
dre:336168 eef2l2, fj53d02, si:ch211-113n10.4, wu:fj53d02; euk... 63.2 2e-10
sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involv... 62.4 3e-10
xla:100505433 hypothetical protein LOC100505433; K03234 elonga... 60.8 9e-10
mmu:13629 Eef2, Ef-2, MGC98463; eukaryotic translation elongat... 60.8 9e-10
hsa:1938 EEF2, EEF-2, EF2; eukaryotic translation elongation f... 60.8 1e-09
dre:326929 eef2b, EEF2, MGC63584, eef2l, fe49h02, wu:fe49h02, ... 59.7 2e-09
xla:380348 eef2.1, MGC53560, eef-2, eef2, ef2; eukaryotic tran... 59.7 2e-09
xla:446712 eef2.2, MGC84492, eef-2, ef2, eft-2; eukaryotic tra... 59.7 2e-09
cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116 k... 57.8 8e-09
dre:568741 Elongation FacTor family member (eft-2)-like; K1453... 56.2 2e-08
cel:F25H5.4 eft-2; Elongation FacTor family member (eft-2); K0... 56.2 3e-08
hsa:79631 EFTUD1, FAM42A, FLJ13119, HsT19294, RIA1; elongation... 55.5 5e-08
mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU0... 55.5 5e-08
tpv:TP01_0529 elongation factor 2; K03234 elongation factor 2 54.3
ath:AT1G56070 LOS1; LOS1; copper ion binding / translation elo... 53.1 2e-07
ath:AT3G12915 GTP binding / GTPase; K03234 elongation factor 2 52.8
tgo:TGME49_005470 elongation factor 2, putative ; K03234 elong... 49.3 3e-06
bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234 ... 48.9 3e-06
sce:YOR133W EFT1; Eft1p; K03234 elongation factor 2 48.5
sce:YDR385W EFT2; Eft2p; K03234 elongation factor 2 48.5
pfa:PF14_0486 elongation factor 2; K03234 elongation factor 2 47.8
cpv:cgd8_2930 Eft2p GTpase; translation elongation factor 2 (E... 46.6 2e-05
sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogene... 45.4 4e-05
ath:AT3G22980 elongation factor Tu family protein; K14536 ribo... 43.1 2e-04
dre:568904 eef2a.2, si:dkey-110c1.4; eukaryotic translation el... 43.1 2e-04
dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191; eu... 40.4 0.001
hsa:4363 ABCC1, ABC29, ABCC, DKFZp686N04233, DKFZp781G125, GS-... 30.0 1.9
cpv:cgd6_3960 elongation factor-like protein 30.0 1.9
tgo:TGME49_068710 elongation factor Tu GTP-binding domain-cont... 29.3 3.4
ath:AT5G43730 disease resistance protein (CC-NBS-LRR class), p... 28.1 6.4
cel:F33G12.4 lrr-1; Leucine-Rich Repeat protein family member ... 28.1 6.4
> tgo:TGME49_086080 U5 small nuclear ribonucleoprotein, putative
(EC:2.7.7.4); K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=1008
Score = 114 bits (286), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 55/76 (72%), Positives = 67/76 (88%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLIGG DLSV KTSTIT+++++ EVEIF PL F + PV+KVACEPLQPSELPKM+E LRR
Sbjct 589 VLIGGVDLSVLKTSTITNVDHSEEVEIFSPLLFNSVPVIKVACEPLQPSELPKMLEALRR 648
Query 61 IDKAYPIAQMKVEESG 76
IDK+YPI++ +VEESG
Sbjct 649 IDKSYPISRTRVEESG 664
> dre:393480 eftud2, MGC66214, wu:fi20f05, zgc:66214; elongation
factor Tu GTP binding domain containing 2; K12852 116 kDa
U5 small nuclear ribonucleoprotein component
Length=973
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/76 (57%), Positives = 57/76 (75%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI GCD + KT+TIT N E +IFRPL F T+ V+K+A EP+ PSELPKM++GLR+
Sbjct 551 VLIEGCDQPIVKTATITEPRGNEEAQIFRPLKFNTASVIKIAVEPVNPSELPKMLDGLRK 610
Query 61 IDKAYPIAQMKVEESG 76
++K+YP KVEESG
Sbjct 611 VNKSYPSLTTKVEESG 626
> ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5);
GTP binding / GTPase/ translation elongation factor/ translation
factor, nucleic acid binding; K12852 116 kDa U5 small nuclear
ribonucleoprotein component
Length=987
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 45/76 (59%), Positives = 56/76 (73%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D S+ KT+T+ + + +V IFR L F T PV+K A EPL PSELPKM+EGLR+
Sbjct 564 VLIEGVDASIMKTATLCNASYDEDVYIFRALQFNTLPVVKTATEPLNPSELPKMVEGLRK 623
Query 61 IDKAYPIAQMKVEESG 76
I K+YP+A KVEESG
Sbjct 624 ISKSYPLAITKVEESG 639
> ath:AT5G25230 elongation factor Tu family protein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=973
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/76 (59%), Positives = 56/76 (73%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D S+ KT+T+ + + +V IFR L F T PV+K A EPL PSELPKM+EGLR+
Sbjct 550 VLIEGVDASIMKTATLCNASYDEDVYIFRALKFNTLPVVKTATEPLNPSELPKMVEGLRK 609
Query 61 IDKAYPIAQMKVEESG 76
I K+YP+A KVEESG
Sbjct 610 ISKSYPLAITKVEESG 625
> xla:379223 eftud2, MGC53479, snrp116, snu114; elongation factor
Tu GTP binding domain containing 2; K12852 116 kDa U5 small
nuclear ribonucleoprotein component
Length=974
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/76 (56%), Positives = 56/76 (73%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D + KT+TIT N E +IFRPL F T+ V+K+A EP+ PSELPKM++GLR+
Sbjct 552 VLIEGVDQPIVKTATITEPRGNEEAQIFRPLKFNTTSVIKIAVEPVNPSELPKMLDGLRK 611
Query 61 IDKAYPIAQMKVEESG 76
++K+YP KVEESG
Sbjct 612 VNKSYPSLTTKVEESG 627
> hsa:9343 EFTUD2, DKFZp686E24196, FLJ44695, KIAA0031, Snrp116,
Snu114, U5-116KD; elongation factor Tu GTP binding domain
containing 2; K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=937
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/76 (56%), Positives = 56/76 (73%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D + KT+TIT N E +IFRPL F T+ V+K+A EP+ PSELPKM++GLR+
Sbjct 515 VLIEGVDQPIVKTATITEPRGNEEAQIFRPLKFNTTSVIKIAVEPVNPSELPKMLDGLRK 574
Query 61 IDKAYPIAQMKVEESG 76
++K+YP KVEESG
Sbjct 575 VNKSYPSLTTKVEESG 590
> mmu:20624 Eftud2, 116kDa, Snrp116, U5-116kD; elongation factor
Tu GTP binding domain containing 2; K12852 116 kDa U5 small
nuclear ribonucleoprotein component
Length=972
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/76 (56%), Positives = 56/76 (73%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D + KT+TIT N E +IFRPL F T+ V+K+A EP+ PSELPKM++GLR+
Sbjct 550 VLIEGVDQPIVKTATITEPRGNEEAQIFRPLKFNTTSVIKIAVEPVNPSELPKMLDGLRK 609
Query 61 IDKAYPIAQMKVEESG 76
++K+YP KVEESG
Sbjct 610 VNKSYPSLTTKVEESG 625
> xla:379973 snrp116-pending, MGC52678; U5 snRNP-specific protein,
116 kD; K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=974
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/76 (56%), Positives = 56/76 (73%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D + KT+TIT N E +IFRPL F T+ V+K+A EP+ PSELPKM++GLR+
Sbjct 552 VLIEGVDQPIVKTATITEPRGNEEAQIFRPLKFNTTSVIKIAVEPVNPSELPKMLDGLRK 611
Query 61 IDKAYPIAQMKVEESG 76
++K+YP KVEESG
Sbjct 612 VNKSYPSLTTKVEESG 627
> bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear protein;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=999
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 44/78 (56%), Positives = 53/78 (67%), Gaps = 2/78 (2%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFR--PLSFYTSPVLKVACEPLQPSELPKMIEGL 58
VLI G DL K TITSL++ EIFR + PV KVA EPL PSELP+M+EGL
Sbjct 578 VLISGIDLCTHKVMTITSLDDPYSAEIFRMSDTLLASEPVFKVAIEPLNPSELPRMVEGL 637
Query 59 RRIDKAYPIAQMKVEESG 76
RRID++YP + +VEESG
Sbjct 638 RRIDRSYPAIKTRVEESG 655
> cel:ZK328.2 eft-1; Elongation FacTor family member (eft-1);
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=974
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/76 (52%), Positives = 54/76 (71%), Gaps = 0/76 (0%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
VLI G D + KT+TI L +V IFRPL F T +K+A EP+ PSELPKM++GLR+
Sbjct 554 VLIEGIDQPIVKTATIAELGYEEDVYIFRPLKFNTRSCVKLAVEPINPSELPKMLDGLRK 613
Query 61 IDKAYPIAQMKVEESG 76
++K+YP+ +VEESG
Sbjct 614 VNKSYPLLTTRVEESG 629
> pfa:PF10_0041 U5 small nuclear ribonuclear protein, putative;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1235
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 36/64 (56%), Positives = 43/64 (67%), Gaps = 6/64 (9%)
Query 19 LENNVEVEIFRPLSFY------TSPVLKVACEPLQPSELPKMIEGLRRIDKAYPIAQMKV 72
L +N EIF PL + V KVACEP+ PSELPKM+EGLR+IDK YP++ KV
Sbjct 744 LNDNENAEIFYPLHKKFRYLNCVNSVFKVACEPINPSELPKMLEGLRKIDKVYPLSSTKV 803
Query 73 EESG 76
EESG
Sbjct 804 EESG 807
> tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=1028
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 52/78 (66%), Gaps = 3/78 (3%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFY--TSPVLKVACEPLQPSELPKMIEGL 58
V++ G D+S KT+T+T N VE+ R S+ PV KV EPL P+ELPKM+ GL
Sbjct 608 VMLSGIDISHYKTTTVTE-NTNSTVELMRIASYLPCVRPVFKVGLEPLNPNELPKMVNGL 666
Query 59 RRIDKAYPIAQMKVEESG 76
R I+K+YP + +KVEESG
Sbjct 667 RSIEKSYPGSLVKVEESG 684
> dre:336168 eef2l2, fj53d02, si:ch211-113n10.4, wu:fj53d02; eukaryotic
translation elongation factor 2, like 2; K03234 elongation
factor 2
Length=861
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 48/72 (66%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ EN+ + R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 477 GVDQFLVKTGTITTFENSHNM---RVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 533
Query 65 YPIAQMKVEESG 76
P+ Q +EESG
Sbjct 534 DPMVQCIIEESG 545
> sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involved
in mRNA splicing via spliceosome; binds directly to U5
snRNA; proposed to be involved in conformational changes of
the spliceosome; similarity to ribosomal translocation factor
EF-2; K12853 114 kDa U5 small nuclear ribonucleoprotein component
Length=1008
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 50/79 (63%), Gaps = 3/79 (3%)
Query 1 VLIGGCDLSVKKTSTITSL---ENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEG 57
VLI G + K++T+ S+ E+ +++ F+PL + T V K+ +PL P ELPK+++
Sbjct 562 VLIKGISSAYIKSATLYSVKSKEDMKQLKFFKPLDYITEAVFKIVLQPLLPRELPKLLDA 621
Query 58 LRRIDKAYPIAQMKVEESG 76
L +I K YP +KVEESG
Sbjct 622 LNKISKYYPGVIIKVEESG 640
> xla:100505433 hypothetical protein LOC100505433; K03234 elongation
factor 2
Length=858
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ E+ R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 474 GVDQFLVKTGTITTFEH---AHNMRVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 530
Query 65 YPIAQMKVEESG 76
P+ Q +EESG
Sbjct 531 DPMVQCIIEESG 542
> mmu:13629 Eef2, Ef-2, MGC98463; eukaryotic translation elongation
factor 2; K03234 elongation factor 2
Length=858
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ E+ R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 474 GVDQFLVKTGTITTFEH---AHNMRVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 530
Query 65 YPIAQMKVEESG 76
P+ Q +EESG
Sbjct 531 DPMVQCIIEESG 542
> hsa:1938 EEF2, EEF-2, EF2; eukaryotic translation elongation
factor 2; K03234 elongation factor 2
Length=858
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ E+ R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 474 GVDQFLVKTGTITTFEH---AHNMRVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 530
Query 65 YPIAQMKVEESG 76
P+ Q +EESG
Sbjct 531 DPMVQCIIEESG 542
> dre:326929 eef2b, EEF2, MGC63584, eef2l, fe49h02, wu:fe49h02,
zgc:63584; eukaryotic translation elongation factor 2b; K03234
elongation factor 2
Length=858
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/72 (41%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ + + R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 474 GVDQFLVKTGTITTFD---QAHNMRVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 530
Query 65 YPIAQMKVEESG 76
P+ Q +EESG
Sbjct 531 DPMVQCIIEESG 542
> xla:380348 eef2.1, MGC53560, eef-2, eef2, ef2; eukaryotic translation
elongation factor 2, gene 1; K03234 elongation factor
2
Length=858
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/72 (41%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TI++ E+ R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 474 GVDQYLVKTGTISTFEH---AHNMRVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 530
Query 65 YPIAQMKVEESG 76
P+ Q +EESG
Sbjct 531 DPMVQCIIEESG 542
> xla:446712 eef2.2, MGC84492, eef-2, ef2, eft-2; eukaryotic translation
elongation factor 2, gene 2
Length=850
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/72 (41%), Positives = 44/72 (61%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 466 GVDQFIVKTGTITTFAG---AHNMRQMKFSVSPVVRVAVECQNPADLPKLVEGLKRLAKS 522
Query 65 YPIAQMKVEESG 76
P+ Q+ EESG
Sbjct 523 DPMVQITTEESG 534
> cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116
kDa ; K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1035
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 33/79 (41%), Positives = 45/79 (56%), Gaps = 3/79 (3%)
Query 1 VLIGGCDLSVKKTSTI---TSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEG 57
VLI G SV K T+ S + E+ R + V+K+A EP P++LPKM+EG
Sbjct 613 VLISGLGSSVTKPCTLIGHNSFIKDDEIYPLRNIRLLNKSVIKLALEPHNPADLPKMLEG 672
Query 58 LRRIDKAYPIAQMKVEESG 76
L+ I KAY + KVEE+G
Sbjct 673 LKSISKAYTCSVTKVEENG 691
> dre:568741 Elongation FacTor family member (eft-2)-like; K14536
ribosome assembly protein 1 [EC:3.6.5.-]
Length=1115
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 51/74 (68%), Gaps = 3/74 (4%)
Query 3 IGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRID 62
IGG + + K++TI++ + F PL+F +P+++VA EP PSE+PK++ G++ ++
Sbjct 586 IGGLEDVILKSATIST---SPACPPFTPLNFEATPIVRVAVEPKHPSEMPKLVRGMKLLN 642
Query 63 KAYPIAQMKVEESG 76
+A P A++ ++E+G
Sbjct 643 QADPCAEVLIQETG 656
> cel:F25H5.4 eft-2; Elongation FacTor family member (eft-2);
K03234 elongation factor 2
Length=852
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 44/72 (61%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + K TIT+ + + R + F SPV++VA E P++LPK++EGL+R+ K+
Sbjct 468 GVDQYLVKGGTITTYK---DAHNMRVMKFSVSPVVRVAVEAKNPADLPKLVEGLKRLAKS 524
Query 65 YPIAQMKVEESG 76
P+ Q EESG
Sbjct 525 DPMVQCIFEESG 536
> hsa:79631 EFTUD1, FAM42A, FLJ13119, HsT19294, RIA1; elongation
factor Tu GTP binding domain containing 1; K14536 ribosome
assembly protein 1 [EC:3.6.5.-]
Length=1069
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 50/74 (67%), Gaps = 3/74 (4%)
Query 3 IGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRID 62
IGG V K++T+ SL + F PL+F +P+++VA EP PSE+P++++G++ ++
Sbjct 534 IGGLQDFVLKSATLCSLPS---CPPFIPLNFEATPIVRVAVEPKHPSEMPQLVKGMKLLN 590
Query 63 KAYPIAQMKVEESG 76
+A P Q+ ++E+G
Sbjct 591 QADPCVQILIQETG 604
> mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU019507,
AU022896, D7Ertd791e; elongation factor Tu GTP binding
domain containing 1; K14536 ribosome assembly protein 1
[EC:3.6.5.-]
Length=1127
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 50/74 (67%), Gaps = 3/74 (4%)
Query 3 IGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRID 62
IGG V K++T+ SL + F PL+F +P+++VA EP PSE+P++++G++ ++
Sbjct 585 IGGLQDFVLKSATLCSLPS---CPPFIPLNFEATPIVRVAVEPKHPSEMPQLVKGMKLLN 641
Query 63 KAYPIAQMKVEESG 76
+A P Q+ ++E+G
Sbjct 642 QADPCVQVLIQETG 655
> tpv:TP01_0529 elongation factor 2; K03234 elongation factor
2
Length=825
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + K+ TIT+ EN + + + SPV++VA +P ELPK++EGL+++ K+
Sbjct 441 GVDQYILKSGTITTFENAYNIA---DMKYSVSPVVRVAVKPKDSKELPKLVEGLKKLSKS 497
Query 65 YPIAQMKVEESG 76
P+ EESG
Sbjct 498 DPLVLCTTEESG 509
> ath:AT1G56070 LOS1; LOS1; copper ion binding / translation elongation
factor/ translation factor, nucleic acid binding;
K03234 elongation factor 2
Length=843
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 47/76 (61%), Gaps = 1/76 (1%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
V + G D + K +T+T+ E V+ R + F SPV++VA + S+LPK++EGL+R
Sbjct 452 VAMVGLDQFITKNATLTN-EKEVDAHPIRAMKFSVSPVVRVAVQCKVASDLPKLVEGLKR 510
Query 61 IDKAYPIAQMKVEESG 76
+ K+ P+ +EESG
Sbjct 511 LAKSDPMVVCTMEESG 526
> ath:AT3G12915 GTP binding / GTPase; K03234 elongation factor
2
Length=820
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 46/76 (60%), Gaps = 1/76 (1%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
V + G D + K T+T+ E V+ R + F SPV++VA + S+LPK++EGL+R
Sbjct 430 VAMVGLDQFITKNGTLTN-EKEVDAHPLRAMKFSVSPVVRVAVKCKLASDLPKLVEGLKR 488
Query 61 IDKAYPIAQMKVEESG 76
+ K+ P+ +EESG
Sbjct 489 LAKSDPMVLCTMEESG 504
> tgo:TGME49_005470 elongation factor 2, putative ; K03234 elongation
factor 2
Length=832
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + K+ T+T+L+ + + + SPV++VA +P ELPK++EGL+++ K+
Sbjct 448 GVDQYLLKSGTLTTLDTAHNIA---DMKYSVSPVVRVAVKPKDNKELPKLVEGLKKLSKS 504
Query 65 YPIAQMKVEESG 76
P+ EESG
Sbjct 505 DPLVVCTTEESG 516
> bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234
elongation factor 2
Length=833
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + K+ TIT+ E + + + SPV++VA +P +LPK++EGL+++ K+
Sbjct 449 GVDQYILKSGTITTCETAHNIA---DMKYSVSPVVRVAVKPKDSKDLPKLVEGLKKLSKS 505
Query 65 YPIAQMKVEESG 76
P+ EESG
Sbjct 506 DPLVVCTTEESG 517
> sce:YOR133W EFT1; Eft1p; K03234 elongation factor 2
Length=842
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT T+T+ E +++ + F SPV++VA E ++LPK++EGL+R+ K+
Sbjct 458 GIDQFLLKTGTLTTSETAHNMKVMK---FSVSPVVQVAVEVKNANDLPKLVEGLKRLSKS 514
Query 65 YPIAQMKVEESG 76
P + ESG
Sbjct 515 DPCVLTYMSESG 526
> sce:YDR385W EFT2; Eft2p; K03234 elongation factor 2
Length=842
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT T+T+ E +++ + F SPV++VA E ++LPK++EGL+R+ K+
Sbjct 458 GIDQFLLKTGTLTTSETAHNMKVMK---FSVSPVVQVAVEVKNANDLPKLVEGLKRLSKS 514
Query 65 YPIAQMKVEESG 76
P + ESG
Sbjct 515 DPCVLTYMSESG 526
> pfa:PF14_0486 elongation factor 2; K03234 elongation factor
2
Length=832
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + K+ TIT+ + E + + SPV++VA +P +LPK+++GL+++ K+
Sbjct 448 GVDQYIVKSGTITTFK---EAHNIADMKYSVSPVVRVAVKPKDSKQLPKLVDGLKKLAKS 504
Query 65 YPIAQMKVEESG 76
P+ +ESG
Sbjct 505 DPLVLCTTDESG 516
> cpv:cgd8_2930 Eft2p GTpase; translation elongation factor 2
(EF-2) ; K03234 elongation factor 2
Length=836
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + K+ TIT+ E + + + SPV++VA P ELPK++EGL+++ K+
Sbjct 452 GIDQYLLKSGTITTSETAHNIA---SMKYSVSPVVRVAVRPKDNKELPKLVEGLKKLSKS 508
Query 65 YPIAQMKVEESG 76
P+ EE+G
Sbjct 509 DPLVVCSKEETG 520
> sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogenesis
of the 60S ribosome; has similarity to translation elongation
factor 2 (Eft1p and Eft2p); K14536 ribosome assembly
protein 1 [EC:3.6.5.-]
Length=1110
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 43/75 (57%), Gaps = 0/75 (0%)
Query 2 LIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRI 61
++G L+ K + T +E V+ ++F+ +P+++VA EP P E+ K++ GL+ +
Sbjct 642 IVGIRGLAGKVLKSGTLIEKGVQGVNLAGVNFHFTPIVRVAVEPANPVEMSKLVRGLKLL 701
Query 62 DKAYPIAQMKVEESG 76
D+A P VE +G
Sbjct 702 DQADPCVHTYVENTG 716
> ath:AT3G22980 elongation factor Tu family protein; K14536 ribosome
assembly protein 1 [EC:3.6.5.-]
Length=1015
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 42/76 (55%), Gaps = 3/76 (3%)
Query 1 VLIGGCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRR 60
V I G + K++T++S N + + F SP L+VA EP P+++ +++GLR
Sbjct 504 VAIRGLGPYISKSATLSSTRNCWPLA---SMEFQVSPTLRVAIEPSDPADMSALMKGLRL 560
Query 61 IDKAYPIAQMKVEESG 76
+++A P ++ V G
Sbjct 561 LNRADPFVEITVSARG 576
> dre:568904 eef2a.2, si:dkey-110c1.4; eukaryotic translation
elongation factor 2a, tandem duplicate 2; K03234 elongation
factor 2
Length=853
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 45/72 (62%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G DL + KT TIT+ + R + F SPV++V+ E + P++LPK++EGL+ + K+
Sbjct 472 GVDLFLVKTGTITTFTKAYNM---RVMKFSVSPVVRVSVEVMDPADLPKLVEGLKHLAKS 528
Query 65 YPIAQMKVEESG 76
P+ Q E+SG
Sbjct 529 DPMLQCIFEDSG 540
> dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191;
eukaryotic translation elongation factor 2a, tandem duplicate
1; K03234 elongation factor 2
Length=854
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 45/72 (62%), Gaps = 3/72 (4%)
Query 5 GCDLSVKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA 64
G D + KT TIT+ + R + F SPV++V+ E + P++LPK++EGL+ + K+
Sbjct 472 GVDQFLVKTGTITTFTKAYNM---RVMKFSVSPVVRVSVEVMDPADLPKLVEGLKHLAKS 528
Query 65 YPIAQMKVEESG 76
P+ Q +E++G
Sbjct 529 DPMLQCIIEDTG 540
> hsa:4363 ABCC1, ABC29, ABCC, DKFZp686N04233, DKFZp781G125, GS-X,
MRP, MRP1; ATP-binding cassette, sub-family C (CFTR/MRP),
member 1; K05665 ATP-binding cassette, subfamily C (CFTR/MRP),
member 1
Length=1531
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 9/29 (31%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 6 CDLSVKKTSTITSLENNVEVEIFRPLSFY 34
C L++ ++ +T+L+ + +V++FR ++FY
Sbjct 148 CALAILRSKIMTALKEDAQVDLFRDITFY 176
> cpv:cgd6_3960 elongation factor-like protein
Length=1100
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 36 SPVLKVACEPLQPSELPKMIEGLRRIDKAYPIAQMKVEESG 76
S ++KV+ EP + +LP M+ GL + ++ P ++ ++G
Sbjct 538 SSIIKVSIEPKRIQDLPLMLRGLELLSRSDPCIEIDTLDTG 578
> tgo:TGME49_068710 elongation factor Tu GTP-binding domain-containing
protein (EC:2.7.7.4); K03234 elongation factor 2
Length=1697
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 27 IFRPLSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKAYPIAQMKVEESG 76
+ P S +S +++VA EP + + + ++G+ R+ A P ++ V +SG
Sbjct 884 LLTPYSKASSAIVRVAVEPQRLEDAQQFVKGMARLYVADPSIKLSVLDSG 933
> ath:AT5G43730 disease resistance protein (CC-NBS-LRR class),
putative
Length=848
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 10 VKKTSTITSLENNVEVEIFRPLSFYTSPVLKVACEPLQPS 49
VK + + + N++ EI R +S ++ V K+AC P P+
Sbjct 495 VKSGAHVRLIPNDISWEIVRQMSLISTQVEKIACSPNCPN 534
> cel:F33G12.4 lrr-1; Leucine-Rich Repeat protein family member
(lrr-1)
Length=453
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Query 31 LSFYTSPVLKVACEPLQPSELPKMIEGLRRIDKA--YPIAQMKVEES 75
++F T+P + V P L ++IE +R+I K I ++KV+ S
Sbjct 77 ITFQTTPAVMVCISKANPEALAQLIEAIRKIVKGENVDIEKVKVKSS 123
Lambda K H
0.317 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017039696
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40