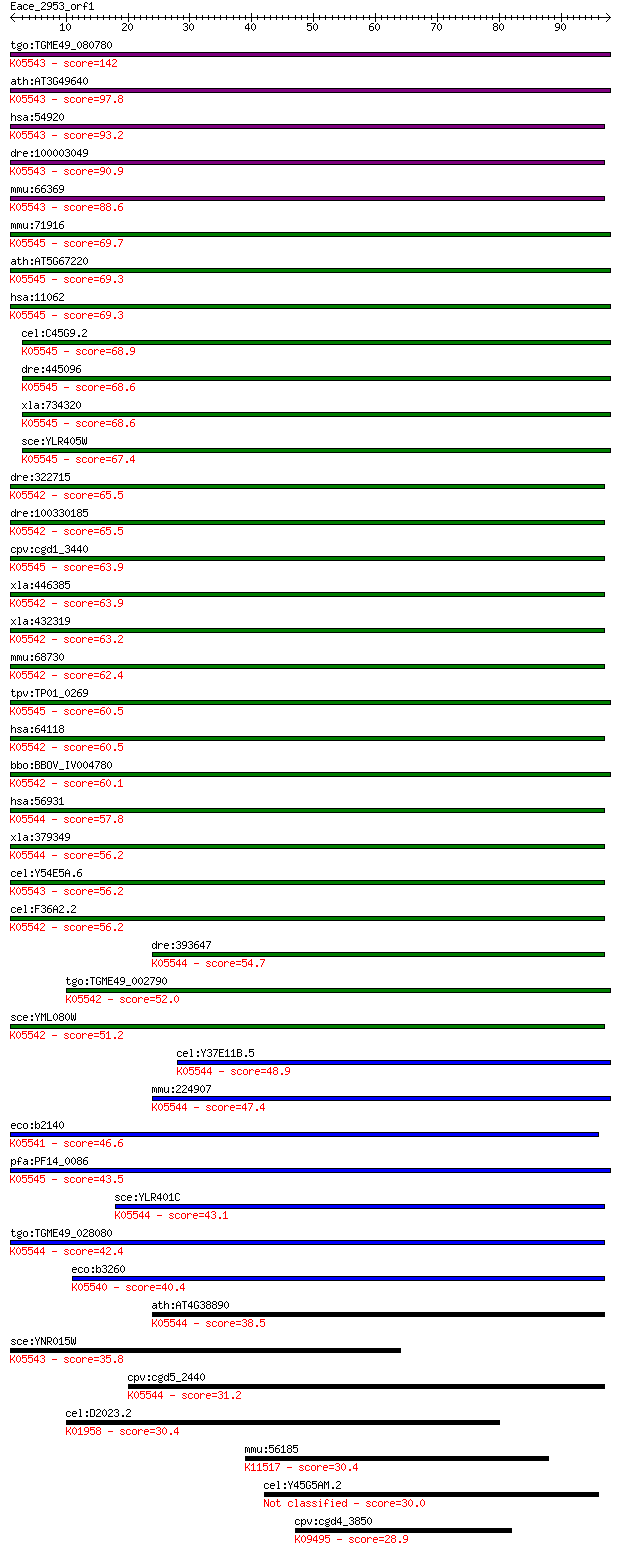
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2953_orf1
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_080780 dihydrouridine synthase domain-containing pr... 142 4e-34
ath:AT3G49640 FAD binding / catalytic/ tRNA dihydrouridine syn... 97.8 7e-21
hsa:54920 DUS2L, DUS2, FLJ20399, SMM1, URLC8; dihydrouridine s... 93.2 2e-19
dre:100003049 dus2l; dihydrouridine synthase 2-like, SMM1 homo... 90.9 1e-18
mmu:66369 Dus2l, 2310016K04Rik; dihydrouridine synthase 2-like... 88.6 5e-18
mmu:71916 Dus4l, 2310069P03Rik, 2700089B10Rik, AI482040, MGC13... 69.7 2e-12
ath:AT5G67220 nitrogen regulation family protein; K05545 tRNA-... 69.3 3e-12
hsa:11062 DUS4L, DUS4, MGC133233, PP35; dihydrouridine synthas... 69.3 3e-12
cel:C45G9.2 hypothetical protein; K05545 tRNA-dihydrouridine s... 68.9 4e-12
dre:445096 dus4l, zgc:92033; dihydrouridine synthase 4-like (S... 68.6 4e-12
xla:734320 dus4l; dihydrouridine synthase 4-like; K05545 tRNA-... 68.6 4e-12
sce:YLR405W DUS4; Dihydrouridine synthase, member of a widespr... 67.4 1e-11
dre:322715 dus1l, wu:fb71f06, zgc:63748; dihydrouridine syntha... 65.5 4e-11
dre:100330185 dihydrouridine synthase 1-like (S. cerevisiae)-l... 65.5 4e-11
cpv:cgd1_3440 Dus1p, tRNA dihydrouridine synthase ; K05545 tRN... 63.9 1e-10
xla:446385 dus1l, MGC83715; dihydrouridine synthase 1-like; K0... 63.9 1e-10
xla:432319 MGC132093; hypothetical protein MGC78973; K05542 tR... 63.2 2e-10
mmu:68730 Dus1l, 1110032N12Rik; dihydrouridine synthase 1-like... 62.4 4e-10
tpv:TP01_0269 hypothetical protein; K05545 tRNA-dihydrouridine... 60.5 1e-09
hsa:64118 DUS1L, DUS1, PP3111; dihydrouridine synthase 1-like ... 60.5 1e-09
bbo:BBOV_IV004780 23.m05837; dihydrouridine synthase; K05542 t... 60.1 2e-09
hsa:56931 DUS3L, DUS3, FLJ13896; dihydrouridine synthase 3-lik... 57.8 9e-09
xla:379349 dus3l, MGC53781; dihydrouridine synthase 3-like; K0... 56.2 2e-08
cel:Y54E5A.6 hypothetical protein; K05543 tRNA-dihydrouridine ... 56.2 3e-08
cel:F36A2.2 hypothetical protein; K05542 tRNA-dihydrouridine s... 56.2 3e-08
dre:393647 MGC63779; zgc:63779; K05544 tRNA-dihydrouridine syn... 54.7 7e-08
tgo:TGME49_002790 dihydrouridine synthase domain-containing pr... 52.0 5e-07
sce:YML080W DUS1; Dihydrouridine synthase, member of a widespr... 51.2 9e-07
cel:Y37E11B.5 hypothetical protein; K05544 tRNA-dihydrouridine... 48.9 3e-06
mmu:224907 Dus3l, AI662135, AW557805, MGC56820; dihydrouridine... 47.4 1e-05
eco:b2140 dusC, ECK2133, JW2128, yohI; tRNA-dihydrouridine syn... 46.6 2e-05
pfa:PF14_0086 tRNA-dihydrouridine synthase, putative; K05545 t... 43.5 2e-04
sce:YLR401C DUS3; Dihydrouridine synthase, member of a widespr... 43.1 2e-04
tgo:TGME49_028080 hypothetical protein ; K05544 tRNA-dihydrour... 42.4 4e-04
eco:b3260 dusB, ECK3247, JW3228, yhdG; tRNA-dihydrouridine syn... 40.4 0.001
ath:AT4G38890 dihydrouridine synthase family protein; K05544 t... 38.5 0.005
sce:YNR015W SMM1, DUS2; Dihydrouridine synthase, member of a f... 35.8 0.036
cpv:cgd5_2440 Ylr401cp-like protein with 2 CCCH domains plus D... 31.2 0.77
cel:D2023.2 pyc-1; PYruvate Carboxylase family member (pyc-1);... 30.4 1.5
mmu:56185 Hao2, AI325478, Hao-2, Hao3, Haox3; hydroxyacid oxid... 30.4 1.5
cel:Y45G5AM.2 hypothetical protein 30.0 2.1
cpv:cgd4_3850 t-complex protein 1, gamma subunit ; K09495 T-co... 28.9 3.8
> tgo:TGME49_080780 dihydrouridine synthase domain-containing
protein ; K05543 tRNA-dihydrouridine synthase 2 [EC:1.-.-.-]
Length=653
Score = 142 bits (357), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 64/97 (65%), Positives = 78/97 (80%), Gaps = 0/97 (0%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLI 60
TCKIRLL+T+ +T+DFAR CE+ GI AIA+HAR+ HERP+HRA+W FP L+S+L IPLI
Sbjct 396 TCKIRLLDTIHKTIDFARLCENCGIEAIAIHARETHERPSHRAHWDLFPALKSSLAIPLI 455
Query 61 ANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
ANGD + DA FQE G SLMFARGAMWDPS+F+
Sbjct 456 ANGDFLTRRDADVFQEKIGADSLMFARGAMWDPSIFS 492
> ath:AT3G49640 FAD binding / catalytic/ tRNA dihydrouridine synthase;
K05543 tRNA-dihydrouridine synthase 2 [EC:1.-.-.-]
Length=290
Score = 97.8 bits (242), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 47/97 (48%), Positives = 64/97 (65%), Gaps = 0/97 (0%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLI 60
TCKIRLL++ +TV+ AR+ E G+ A+AVH RK +RP A W + +AL IP+I
Sbjct 108 TCKIRLLKSPADTVELARRIEKLGVPALAVHGRKIADRPRDPAKWDEIADVVAALSIPVI 167
Query 61 ANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
ANGD+ DD R + A G S+M ARGAMW+ S+F+
Sbjct 168 ANGDVLEYDDFSRIKTATGAASVMVARGAMWNASIFS 204
> hsa:54920 DUS2L, DUS2, FLJ20399, SMM1, URLC8; dihydrouridine
synthase 2-like, SMM1 homolog (S. cerevisiae); K05543 tRNA-dihydrouridine
synthase 2 [EC:1.-.-.-]
Length=493
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/100 (46%), Positives = 62/100 (62%), Gaps = 4/100 (4%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLI 60
TCKIR+L +LE+T+ ++ E TGIAAIAVH RKR ERP H + + + L IP+I
Sbjct 153 TCKIRILPSLEDTLSLVKRIERTGIAAIAVHGRKREERPQHPVSCEVIKAIADTLSIPVI 212
Query 61 ANGD----LFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
ANG + D F++A S+M AR AMW+PS+F
Sbjct 213 ANGGSHDHIQQYSDIEDFRQATAASSVMVARAAMWNPSIF 252
> dre:100003049 dus2l; dihydrouridine synthase 2-like, SMM1 homolog
(S. cerevisiae); K05543 tRNA-dihydrouridine synthase 2
[EC:1.-.-.-]
Length=504
Score = 90.9 bits (224), Expect = 1e-18, Method: Composition-based stats.
Identities = 44/100 (44%), Positives = 65/100 (65%), Gaps = 4/100 (4%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLI 60
TCKIR+L T+E+TV+ ++ E TG+AAIAVH R + ERP H + + ++ +P+I
Sbjct 158 TCKIRILSTIEDTVNLVKRIEKTGVAAIAVHGRMKDERPRHPVHCDYIQAVAQSVSVPVI 217
Query 61 ANG---DLFSA-DDAIRFQEAYGIRSLMFARGAMWDPSLF 96
ANG D+ +D F+E+ G S+M AR AMW+PS+F
Sbjct 218 ANGGSSDIVKEFEDIETFRESTGASSVMLARAAMWNPSVF 257
> mmu:66369 Dus2l, 2310016K04Rik; dihydrouridine synthase 2-like
(SMM1, S. cerevisiae); K05543 tRNA-dihydrouridine synthase
2 [EC:1.-.-.-]
Length=493
Score = 88.6 bits (218), Expect = 5e-18, Method: Composition-based stats.
Identities = 45/103 (43%), Positives = 64/103 (62%), Gaps = 10/103 (9%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLI 60
TCKIR+L +LE+T++ ++ E TGI+AIAVH R R ERP H + + + L IP+I
Sbjct 153 TCKIRILPSLEDTLNLVKRIERTGISAIAVHGRNRDERPQHPVSCEVIRAIAETLSIPVI 212
Query 61 ANGDLFSADDAIR-------FQEAYGIRSLMFARGAMWDPSLF 96
ANG + D I+ F++A S+M AR AMW+PS+F
Sbjct 213 ANG---GSHDHIQQHVDIEDFRQATAASSVMVARAAMWNPSIF 252
> mmu:71916 Dus4l, 2310069P03Rik, 2700089B10Rik, AI482040, MGC130363,
Pp35; dihydrouridine synthase 4-like (S. cerevisiae);
K05545 tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=324
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 36/99 (36%), Positives = 58/99 (58%), Gaps = 5/99 (5%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHER--PAHRANWQAFPVLQSALRIP 58
+ KIR+ + L T+D RK E+TG++ I VH R ER P H + A +++ + IP
Sbjct 156 SIKIRIHDDLARTIDLCRKAEATGVSWITVHGRTVEERHQPVH---YDAIKMIKENVSIP 212
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
++ANGD+ S +A + G +M ARG + +P++FA
Sbjct 213 IVANGDIRSLKEAENVWQMTGTDGVMVARGLLTNPAMFA 251
> ath:AT5G67220 nitrogen regulation family protein; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=423
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 59/99 (59%), Gaps = 2/99 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAH--RANWQAFPVLQSALRIP 58
+CKIR+ LE+T+ +A+ E G + +AVH R R E+ RA+W A +++A+RIP
Sbjct 214 SCKIRIFPNLEDTLKYAKMLEDAGCSLLAVHGRTRDEKDGKKFRADWSAIKEVKNAMRIP 273
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
++ANG++ +D + G+ ++ A + +P+ FA
Sbjct 274 VLANGNVRCIEDVDNCIKETGVEGVLSAETLLENPAAFA 312
> hsa:11062 DUS4L, DUS4, MGC133233, PP35; dihydrouridine synthase
4-like (S. cerevisiae); K05545 tRNA-dihydrouridine synthase
4 [EC:1.-.-.-]
Length=317
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 36/99 (36%), Positives = 58/99 (58%), Gaps = 5/99 (5%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHER--PAHRANWQAFPVLQSALRIP 58
+ KIR+ + L+ TVD +K E+TG++ I VH R ER P H + + +++ + IP
Sbjct 156 SIKIRIHDDLKRTVDLCQKAEATGVSWITVHGRTAEERHQPVH---YDSIKIIKENMSIP 212
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
+IANGD+ S +A G +M ARG + +P++FA
Sbjct 213 VIANGDIRSLKEAENVWRITGTDGVMVARGLLANPAMFA 251
> cel:C45G9.2 hypothetical protein; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=284
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 58/95 (61%), Gaps = 1/95 (1%)
Query 3 KIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLIAN 62
KIR+ +E+TVD RK E+ G+ + VH R +R A + QA +++ ++ +P+IAN
Sbjct 125 KIRINHDIEKTVDLCRKAEAAGVTHLTVHGRTPSQR-AEPIDIQALRIVKDSVSVPIIAN 183
Query 63 GDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
G + + ++A+ E G+ +M A G + +P+LFA
Sbjct 184 GGITTREEALFLAEQTGVDGIMAANGLLDNPALFA 218
> dre:445096 dus4l, zgc:92033; dihydrouridine synthase 4-like
(S. cerevisiae); K05545 tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=285
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 56/97 (57%), Gaps = 5/97 (5%)
Query 3 KIRLLETLEETVDFARKCESTGIAAIAVHARKRHER--PAHRANWQAFPVLQSALRIPLI 60
KIR+ + + +TVD +K E+ G++ I VH R ER P H + ++ +L IP+I
Sbjct 125 KIRIHKDVRQTVDLCQKVEAAGVSYITVHGRTAIERHQPVH---YDTIKTIKDSLSIPVI 181
Query 61 ANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
ANGD+ S D E G+ +M ARG + +P++F+
Sbjct 182 ANGDIKSMQDVEAVCELTGVDGVMSARGLLSNPAMFS 218
> xla:734320 dus4l; dihydrouridine synthase 4-like; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=308
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 5/97 (5%)
Query 3 KIRLLETLEETVDFARKCESTGIAAIAVHARKRHER--PAHRANWQAFPVLQSALRIPLI 60
KIR+ + TVD RK E+ G++ I VH R ER P H + A ++ +L IP++
Sbjct 148 KIRIHADISRTVDLCRKAEAAGVSWITVHGRTHDERHQPVH---YNAIKNIKESLSIPIV 204
Query 61 ANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
ANGD+ S +A + G +M ARG + +P++FA
Sbjct 205 ANGDIKSLKEAENIHQITGADGVMAARGLLANPAMFA 241
> sce:YLR405W DUS4; Dihydrouridine synthase, member of a widespread
family of conserved proteins including Smm1p, Dus1p, and
Dus3p (EC:1.-.-.-); K05545 tRNA-dihydrouridine synthase 4
[EC:1.-.-.-]
Length=367
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 36/98 (36%), Positives = 51/98 (52%), Gaps = 3/98 (3%)
Query 3 KIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSAL---RIPL 59
KIR+ E L+ETV+ RK G+ I +H R R R + AN A + + +P+
Sbjct 169 KIRIHEALDETVELCRKLCDAGVDWITIHGRTRRTRSSQPANLDAIKYIIENISDKNVPV 228
Query 60 IANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
IANGD F D R + G +M RG + +P+LFA
Sbjct 229 IANGDCFKLSDLERITKYTGAHGVMAVRGLLSNPALFA 266
> dre:322715 dus1l, wu:fb71f06, zgc:63748; dihydrouridine synthase
1-like (S. cerevisiae); K05542 tRNA-dihydrouridine synthase
1 [EC:1.-.-.-]
Length=479
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 32/98 (32%), Positives = 55/98 (56%), Gaps = 2/98 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSALRIP 58
TCKIR+ +E+TV +A+ E G + VH R + ++ A A+W+ ++ A+ IP
Sbjct 145 TCKIRVFPEIEKTVKYAKMLEKAGCQLLTVHGRTKDQKGALTGIASWKHIKAVREAVNIP 204
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+ ANG++ D E G++ +M A G + +P+LF
Sbjct 205 VFANGNIQHLSDVHHCMEETGVQGVMSAEGNLHNPALF 242
> dre:100330185 dihydrouridine synthase 1-like (S. cerevisiae)-like;
K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=496
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 32/98 (32%), Positives = 55/98 (56%), Gaps = 2/98 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSALRIP 58
TCKIR+ +E+TV +A+ E G + VH R + ++ A A+W+ ++ A+ IP
Sbjct 162 TCKIRVFPEIEKTVKYAKMLEKAGCQLLTVHGRTKDQKGALTGIASWKHIKAVREAVNIP 221
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+ ANG++ D E G++ +M A G + +P+LF
Sbjct 222 VFANGNIQHLSDVHHCMEETGVQGVMSAEGNLHNPALF 259
> cpv:cgd1_3440 Dus1p, tRNA dihydrouridine synthase ; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=409
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 36/100 (36%), Positives = 55/100 (55%), Gaps = 4/100 (4%)
Query 1 TCKIRLLE--TLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSALR 56
+CKIR+L+ L+ T++ ++ ES G IAVH R R ANW+A +L+S
Sbjct 146 SCKIRILDHHDLQPTINLIKRLESAGACMIAVHGRTMTSRGVLTGPANWEALKILKSRCS 205
Query 57 IPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
IP IANG + + +D + G ++M A G + +P LF
Sbjct 206 IPFIANGGISNYEDIQKCLNYTGADAVMSAEGILENPWLF 245
> xla:446385 dus1l, MGC83715; dihydrouridine synthase 1-like;
K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=464
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 56/98 (57%), Gaps = 2/98 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSALRIP 58
TCKIR+ +E+TV +A+ E G + VH R + ++ + A+W+ +++A+ IP
Sbjct 139 TCKIRVFPEMEKTVQYAKMLEKAGCQLLTVHGRTKEQKGSLTGIASWEHIKAVRNAVNIP 198
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+ ANG++ +D R ++ +M A G + +P+LF
Sbjct 199 VFANGNIQYLNDVERCMRETAVQGVMSAEGNLHNPALF 236
> xla:432319 MGC132093; hypothetical protein MGC78973; K05542
tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=465
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 56/98 (57%), Gaps = 2/98 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSALRIP 58
TCKIR+ +E+TV++A+ E G + VH R + ++ A+W+ +++A+ IP
Sbjct 139 TCKIRVFPEIEKTVEYAKMLEKAGCQLLTVHGRTKEQKGPLTGIASWEHIKAVRNAVIIP 198
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+ ANG++ +D R ++ +M A G + +P+LF
Sbjct 199 VFANGNIQYLNDVERCMRETAVQGVMSAEGNLHNPALF 236
> mmu:68730 Dus1l, 1110032N12Rik; dihydrouridine synthase 1-like
(S. cerevisiae); K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=475
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 31/98 (31%), Positives = 56/98 (57%), Gaps = 2/98 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERP--AHRANWQAFPVLQSALRIP 58
TCKIR+ +++TV +A+ E G + VH R + ++ A A+W+ ++ A+ IP
Sbjct 145 TCKIRVFPEIDKTVRYAQMLEKAGCQLLTVHGRTKEQKGPMAGTASWEHIKAVRKAVGIP 204
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+ ANG++ D R + G++ +M A G + +P+LF
Sbjct 205 VFANGNIQCLQDVERCIQDTGVQGVMSAEGNLHNPALF 242
> tpv:TP01_0269 hypothetical protein; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=327
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 32/101 (31%), Positives = 55/101 (54%), Gaps = 4/101 (3%)
Query 1 TCKIRLLET--LEETVDFARKCESTGIAAIAVHARKRHERPAH--RANWQAFPVLQSALR 56
TCKIR +E L+ T++ E++G A+ VH R R E +W + +++S +
Sbjct 151 TCKIRKVEKDCLQSTLNLCYSLEASGCKALTVHGRHRSETKTRISECDWDSIKIIKSKVN 210
Query 57 IPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
IP+IANG + + +D R E G ++M + + +P LF+
Sbjct 211 IPVIANGGIETFEDVKRCLEYTGADAVMSSEAILENPYLFS 251
> hsa:64118 DUS1L, DUS1, PP3111; dihydrouridine synthase 1-like
(S. cerevisiae); K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=473
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 55/98 (56%), Gaps = 2/98 (2%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERP--AHRANWQAFPVLQSALRIP 58
TCKIR+ +++TV +A+ E G + VH R + ++ + A+W+ ++ A+ IP
Sbjct 145 TCKIRVFPEIDKTVRYAQMLEKAGCQLLTVHGRTKEQKGPLSGAASWEHIKAVRKAVAIP 204
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+ ANG++ D R G++ +M A G + +P+LF
Sbjct 205 VFANGNIQCLQDVERCLRDTGVQGVMSAEGNLHNPALF 242
> bbo:BBOV_IV004780 23.m05837; dihydrouridine synthase; K05542
tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=355
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 34/101 (33%), Positives = 56/101 (55%), Gaps = 4/101 (3%)
Query 1 TCKIRLLE--TLEETVDFARKCESTGIAAIAVHARKRHERPAH--RANWQAFPVLQSALR 56
TCKIR ++ +L+ T++ E +G A+ VH R R E+ + A+W A +++S LR
Sbjct 138 TCKIRKVDKDSLQSTLNLCYSLEQSGCKALTVHGRHRSEKGTNVGAADWDAIRIIKSRLR 197
Query 57 IPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
IP+IANG + + D R G ++M + + P LF+
Sbjct 198 IPVIANGGIETFADVERCLSYTGADAVMSSESLLERPYLFS 238
> hsa:56931 DUS3L, DUS3, FLJ13896; dihydrouridine synthase 3-like
(S. cerevisiae); K05544 tRNA-dihydrouridine synthase 3 [EC:1.-.-.-]
Length=408
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 54/101 (53%), Gaps = 8/101 (7%)
Query 1 TCKIRLLETLEETVDFARKC----ESTGIAAIAVHARKRHERPAHRANWQAF-PVLQSAL 55
T KIR ++E V+ A + G+A + +H R R +R A+WQ +Q+A
Sbjct 191 TVKIR--TGVQERVNLAHRLLPELRDWGVALVTLHGRSREQRYTKLADWQYIEECVQAAS 248
Query 56 RIPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+PL NGD+ S +DA R + G+ +M ARGA+ P LF
Sbjct 249 PMPLFGNGDILSFEDANRAMQT-GVTGIMIARGALLKPWLF 288
> xla:379349 dus3l, MGC53781; dihydrouridine synthase 3-like;
K05544 tRNA-dihydrouridine synthase 3 [EC:1.-.-.-]
Length=640
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 35/101 (34%), Positives = 53/101 (52%), Gaps = 8/101 (7%)
Query 1 TCKIRLLETLEETVDFARKC----ESTGIAAIAVHARKRHERPAHRANWQAF-PVLQSAL 55
T KIR ++E V+ A K G++ + +H R R +R ANW+ + A
Sbjct 423 TVKIR--TGVQEKVNIAHKLIPNLRDWGVSLVTLHGRSREQRYTKLANWEYIDQCAKIAS 480
Query 56 RIPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+PL NGD+ S +DA R + G+ +M ARGA++ P LF
Sbjct 481 PVPLFGNGDIISYEDANRALQT-GVAGVMLARGALFKPWLF 520
> cel:Y54E5A.6 hypothetical protein; K05543 tRNA-dihydrouridine
synthase 2 [EC:1.-.-.-]
Length=436
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/100 (33%), Positives = 53/100 (53%), Gaps = 4/100 (4%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAF-PVLQSALRIPL 59
TCKIR+L+ E+T+ ++ E ++A+ VH R+R ER + V Q+ IP+
Sbjct 147 TCKIRVLDDPEDTLKLVQEIEKCRVSALGVHGRRRDERQPDKCRIDEIRDVAQAVQSIPV 206
Query 60 IANG---DLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
I+NG ++ D ++Q S M AR A+ PS+F
Sbjct 207 ISNGLSDEIEQYSDFEKYQLLTETSSTMIARKALSTPSIF 246
> cel:F36A2.2 hypothetical protein; K05542 tRNA-dihydrouridine
synthase 1 [EC:1.-.-.-]
Length=527
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 56/100 (56%), Gaps = 4/100 (4%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSAL--R 56
+CKIR+ + ++TV++A++ G + VH R R + A A+W + A+ R
Sbjct 199 SCKIRVRDDRQQTVEYAKRLVDAGATMLTVHGRTRDMKGAETGLADWSRIRDVVEAVGSR 258
Query 57 IPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
+P++ANG++ D R +A G ++M A G +++P +F
Sbjct 259 VPVMANGNIQFPGDVERCMQATGAVAIMSAEGLLYNPLIF 298
> dre:393647 MGC63779; zgc:63779; K05544 tRNA-dihydrouridine synthase
3 [EC:1.-.-.-]
Length=660
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 28/74 (37%), Positives = 42/74 (56%), Gaps = 2/74 (2%)
Query 24 GIAAIAVHARKRHERPAHRANWQAFPVL-QSALRIPLIANGDLFSADDAIRFQEAYGIRS 82
G++ I +H R R +R A+W Q A +PL NGD+ S +DA++ +E G+
Sbjct 468 GVSLITLHGRSREQRYTKLADWDYINTCAQLAAPVPLFGNGDILSYEDAMKARET-GVSG 526
Query 83 LMFARGAMWDPSLF 96
+M ARGA+ P LF
Sbjct 527 IMVARGALIKPWLF 540
> tgo:TGME49_002790 dihydrouridine synthase domain-containing
protein (EC:5.1.3.9); K05542 tRNA-dihydrouridine synthase 1
[EC:1.-.-.-]
Length=540
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 10 LEETVDFARKCESTGIAAIAVHARKRHERPAH--RANWQAFPVLQSALRIPLIANGDLFS 67
L++T+ E+ G A + +H R + E+ A +W A ++ L IP+IANG + +
Sbjct 242 LQDTLRLCDAFEAAGCACLCIHGRTKEEKAAFVGPCDWLAIRHVKQRLSIPVIANGAVET 301
Query 68 ADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
+DA+R E G ++M A G + +P LFA
Sbjct 302 YEDALRCLEFTGADAVMSAEGLLDNPMLFA 331
> sce:YML080W DUS1; Dihydrouridine synthase, member of a widespread
family of conserved proteins including Smm1p, Dus3p, and
Dus4p; modifies pre-tRNA(Phe) at U17 (EC:1.-.-.-); K05542
tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=423
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 50/100 (50%), Gaps = 4/100 (4%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARKRHERPAHR--ANWQAFPVLQSAL--R 56
T KIR+ + E+++++A+ G + VH R R ++ ANW+ L+ L
Sbjct 158 TAKIRIFDDCEKSLNYAKMVLDAGAQFLTVHGRVREQKGQKTGLANWETIKYLRDNLPKE 217
Query 57 IPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLF 96
ANG++ +D R E G ++M A G +++P +F
Sbjct 218 TVFFANGNILYPEDISRCMEHIGADAVMSAEGNLYNPGVF 257
> cel:Y37E11B.5 hypothetical protein; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=554
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 35/71 (49%), Gaps = 1/71 (1%)
Query 28 IAVHARKRHERPAHRANWQ-AFPVLQSALRIPLIANGDLFSADDAIRFQEAYGIRSLMFA 86
I H R + +R ANW FPV ++ +PL GD+ S +D E Y + +M
Sbjct 366 ITFHPRSKEQRYTRLANWDFTFPVQEATKPVPLWVCGDILSWEDYYERLEQYPVNGIMIG 425
Query 87 RGAMWDPSLFA 97
RGA+ P +F
Sbjct 426 RGALIKPWIFT 436
> mmu:224907 Dus3l, AI662135, AW557805, MGC56820; dihydrouridine
synthase 3-like (S. cerevisiae); K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=637
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 41/78 (52%), Gaps = 8/78 (10%)
Query 24 GIAAIAVHARKRHERPAHRANWQAFPVLQSALRI----PLIANGDLFSADDAIRFQEAYG 79
G+A + +H R R +R A+W P ++ ++ PL NGD+ S +DA + G
Sbjct 445 GVALVTLHGRSREQRYTRLADW---PYIEQCAKVASPMPLFGNGDILSFEDANCAMQT-G 500
Query 80 IRSLMFARGAMWDPSLFA 97
+ +M ARGA+ P LF
Sbjct 501 VAGIMVARGALLKPWLFT 518
> eco:b2140 dusC, ECK2133, JW2128, yohI; tRNA-dihydrouridine synthase
C; K05541 tRNA-dihydrouridine synthase C [EC:1.-.-.-]
Length=315
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 50/97 (51%), Gaps = 2/97 (2%)
Query 1 TCKIRL-LETLEETVDFARKCESTGIAAIAVHARKRHE-RPAHRANWQAFPVLQSALRIP 58
+ K+RL ++ E+ + A + G + VH R + + A +WQA ++ L IP
Sbjct 137 SVKVRLGWDSGEKKFEIADAVQQAGATELVVHGRTKEQGYRAEHIDWQAIGDIRQRLNIP 196
Query 59 LIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSL 95
+IANG+++ A + G ++M RGA+ P+L
Sbjct 197 VIANGEIWDWQSAQQCMAISGCDAVMIGRGALNIPNL 233
> pfa:PF14_0086 tRNA-dihydrouridine synthase, putative; K05545
tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=340
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 51/100 (51%), Gaps = 3/100 (3%)
Query 1 TCKIRLLET-LEETVDFARKCESTGIAAIAVHARKRHERPAH--RANWQAFPVLQSALRI 57
TCKIR ++ ++T++ +S GI I VH R + E+ + + +++ +++ L I
Sbjct 152 TCKIRKIDNDYQKTLNLCYDLQSRGIKMITVHGRTKEEKGINIKQCDYEIIRIIKERLNI 211
Query 58 PLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSLFA 97
P+IANG + +D + ++M A + P F+
Sbjct 212 PIIANGSIEHFEDIKKCLNYTKADAVMCAEILLEKPYFFS 251
> sce:YLR401C DUS3; Dihydrouridine synthase, member of a widespread
family of conserved proteins including Smm1p, Dus1p, and
Dus4p; contains a consensus oleate response element (ORE)
in its promoter region (EC:1.-.-.-); K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=668
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 44/101 (43%), Gaps = 22/101 (21%)
Query 18 RKCESTGIAAIAVHARKRHERPAHRANW----QAFPVLQSAL-----------------R 56
R T +AAI +H R R +R A+W Q L+SA R
Sbjct 444 RLVNETDVAAITLHGRSRQQRYTKSADWDYVSQVADTLRSAEADFIETEQGKEGRDSKNR 503
Query 57 IPLIANGDLFSADDAIRFQEA-YGIRSLMFARGAMWDPSLF 96
I + NGD+ + +D R+ I S+M ARGA+ P +F
Sbjct 504 IQFVGNGDVNNFEDWYRYLNGNENIDSVMVARGALIKPWIF 544
> tgo:TGME49_028080 hypothetical protein ; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=1220
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 48/101 (47%), Gaps = 6/101 (5%)
Query 1 TCKIRLLETLEETV--DFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSAL--R 56
T K+R ++ V F R+ G AA+ VH R +R A+W + AL
Sbjct 950 TVKLRTANQGKKQVLHSFIRRLGDAGAAALIVHGRTAQQRYTKAADWSYVAQCREALDSS 1009
Query 57 IPLIANGDLFSADDAIRFQEAYGIR-SLMFARGAMWDPSLF 96
PLI GD+FS+ + F+ A G LM AR A+ P +F
Sbjct 1010 TPLIGCGDIFSSPE-FEFRLASGASDGLMIARAALVKPWVF 1049
> eco:b3260 dusB, ECK3247, JW3228, yhdG; tRNA-dihydrouridine synthase
B; K05540 tRNA-dihydrouridine synthase B [EC:1.-.-.-]
Length=321
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 38/86 (44%), Gaps = 3/86 (3%)
Query 11 EETVDFARKCESTGIAAIAVHARKRHERPAHRANWQAFPVLQSALRIPLIANGDLFSADD 70
EE A C GI A+ +H R R A + + ++ + IP+IANGD+
Sbjct 152 EEIAQLAEDC---GIQALTIHGRTRACLFNGEAEYDSIRAVKQKVSIPVIANGDITDPLK 208
Query 71 AIRFQEAYGIRSLMFARGAMWDPSLF 96
A + G +LM R A P +F
Sbjct 209 ARAVLDYTGADALMIGRAAQGRPWIF 234
> ath:AT4G38890 dihydrouridine synthase family protein; K05544
tRNA-dihydrouridine synthase 3 [EC:1.-.-.-]
Length=700
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 24 GIAAIAVHARKRHERPAHRANWQ-AFPVLQSA-LRIPLIANGDLFSADDAIRFQ-EAYGI 80
G A+ +H R R +R + A+W + ++A + +I NGD++S D + + + +
Sbjct 500 GATAVTIHGRSRQQRYSKSADWDYIYQCTKNATTNLQVIGNGDVYSYLDWNKHKSDCPEL 559
Query 81 RSLMFARGAMWDPSLF 96
S M ARGA+ P +F
Sbjct 560 SSCMIARGALIKPWIF 575
> sce:YNR015W SMM1, DUS2; Dihydrouridine synthase, member of a
family of dihydrouridine synthases including Dus1p, Smm1p,
Dus3p, and Dus4p; modifies uridine residues at position 20 of
cytoplasmic tRNAs (EC:1.-.-.-); K05543 tRNA-dihydrouridine
synthase 2 [EC:1.-.-.-]
Length=384
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 36/67 (53%), Gaps = 5/67 (7%)
Query 1 TCKIRLLETLEETVDFARKCESTGIAAIAVHARK----RHERPAHRANWQAFPVLQSALR 56
+ KIRLL+T ++T+ ++ +TGI + VH RK E+P + + + Q A
Sbjct 158 SVKIRLLDTKQDTLQLVKRLCATGITNLTVHCRKTEMRNREQPITDYIAEIYEICQ-ANN 216
Query 57 IPLIANG 63
+ LI NG
Sbjct 217 VSLIVNG 223
> cpv:cgd5_2440 Ylr401cp-like protein with 2 CCCH domains plus
Dus1p tRNA dihydrouridine synthase Tim barrel ; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=707
Score = 31.2 bits (69), Expect = 0.77, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 38/97 (39%), Gaps = 21/97 (21%)
Query 20 CESTGIAAIAVHARKRHERPAHRANWQAF---PVLQSALRIPLIANG-----------DL 65
C+S GI A+ +H R +R A+W VL S L + N +
Sbjct 490 CDS-GIDALTIHGRTSFQRYTKEADWSYIKYCSVLNSKLYSNKVVNNIQNSNHSLICPSI 548
Query 66 FSADDAIRFQEAYG------IRSLMFARGAMWDPSLF 96
D I FQ+ + S+M RGA+ P +F
Sbjct 549 IGCGDIINFQDYQQHIQNDLVDSVMIGRGALLKPWIF 585
> cel:D2023.2 pyc-1; PYruvate Carboxylase family member (pyc-1);
K01958 pyruvate carboxylase [EC:6.4.1.1]
Length=1175
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 10/74 (13%)
Query 10 LEETVDFARKCESTGIAAIA----VHARKRHERPAHRANWQAFPVLQSALRIPLIANGDL 65
L E DFA C++ GI I V AR + A +A +++ +++ G +
Sbjct 116 LSERSDFAAACQNAGIVFIGPSPDVMARMGDKVAARQA------AIEAGVQVVPGTPGPI 169
Query 66 FSADDAIRFQEAYG 79
+AD+A+ F + YG
Sbjct 170 TTADEAVEFAKQYG 183
> mmu:56185 Hao2, AI325478, Hao-2, Hao3, Haox3; hydroxyacid oxidase
2 (EC:1.1.3.15); K11517 (S)-2-hydroxy-acid oxidase [EC:1.1.3.15]
Length=353
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 39 PAHRANWQAFPVLQSALRIPLIANGDLFSADDAIRFQEAYGIRSLMFAR 87
P+ + W P+LQS R+P+I G L D + + + IR ++ +
Sbjct 201 PSSSSCWNDLPLLQSMTRLPIILKGILTKEDAELAVK--HNIRGIIVSN 247
> cel:Y45G5AM.2 hypothetical protein
Length=1061
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 42 RANWQAFPVLQSALRIPLIANGDLFSADDAIRFQEAYGIRSLMFARGAMWDPSL 95
R W ++P + + L+ G + A + YG R +FAR A+WD L
Sbjct 871 RTIWDSYPTEEFMNQYTLVLKGHIRLASASFPKTLTYGSRLDIFARIALWDAGL 924
> cpv:cgd4_3850 t-complex protein 1, gamma subunit ; K09495 T-complex
protein 1 subunit gamma
Length=559
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 47 AFPVLQSALRIPLIANGDLFSADDAIRFQEAYGIR 81
A P+LQ + +IA G L + DD+I+F E ++
Sbjct 110 AEPLLQKNIHPTIIAKGYLRALDDSIKFMEEMSVK 144
Lambda K H
0.326 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40