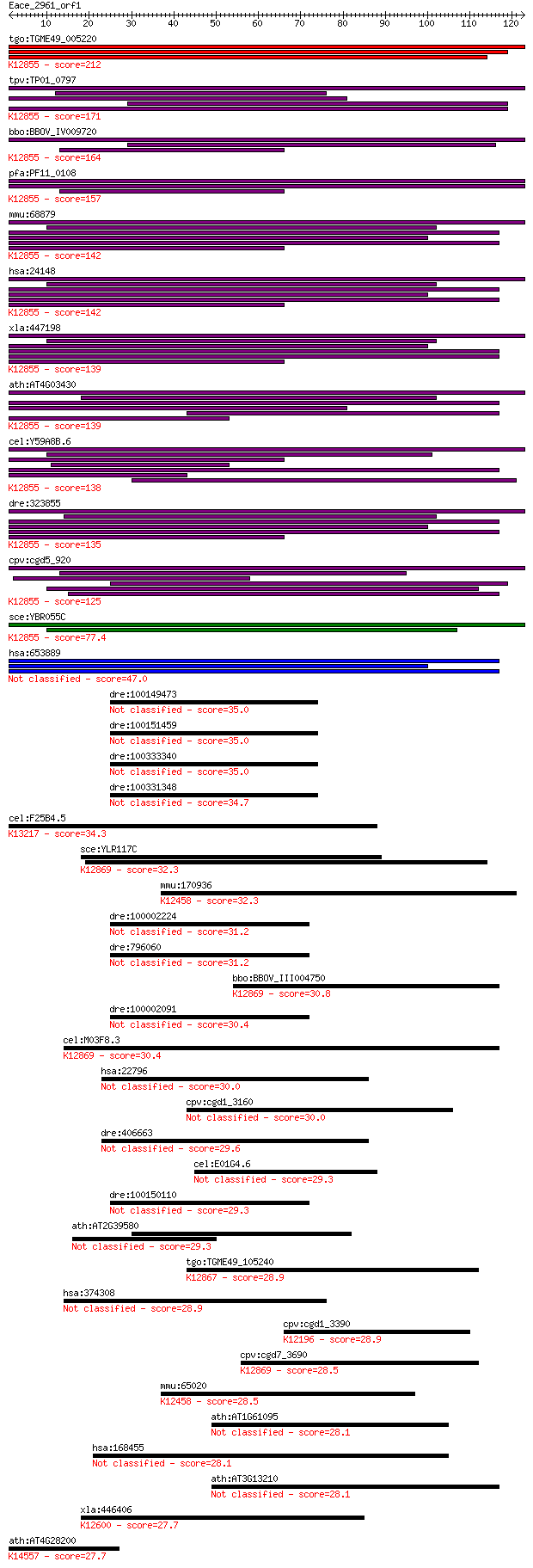
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2961_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putativ... 212 2e-55
tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing... 171 7e-43
bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, puta... 164 5e-41
pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855 pr... 157 5e-39
mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655... 142 3e-34
hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6; P... 142 3e-34
xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor 6 ... 139 2e-33
ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing fa... 139 2e-33
cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing ... 138 4e-33
dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02, z... 135 3e-32
cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat pr... 125 2e-29
sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-proces... 77.4 1e-14
hsa:653889 pre-mRNA-processing factor 6-like 47.0 2e-05
dre:100149473 polyprotein-like 35.0 0.054
dre:100151459 Gag-Pol polyprotein-like 35.0 0.058
dre:100333340 polyprotein-like 35.0 0.063
dre:100331348 polyprotein-like 34.7 0.074
cel:F25B4.5 hypothetical protein; K13217 pre-mRNA-processing f... 34.3 0.086
sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck 32.3 0.38
mmu:170936 Zfp369, B930030B22Rik, D230020H11Rik, NRIF2; zinc f... 32.3 0.38
dre:100002224 polyprotein-like 31.2 0.86
dre:796060 polyprotein-like 31.2 0.87
bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain ... 30.8 1.0
dre:100002091 polyprotein-like 30.4 1.4
cel:M03F8.3 hypothetical protein; K12869 crooked neck 30.4 1.6
hsa:22796 COG2, LDLC; component of oligomeric golgi complex 2 30.0
cpv:cgd1_3160 mRNA 3' end processing protein RNA14, HAT repeats 30.0 1.9
dre:406663 cog2, zC8A9.2, zgc:56436; component of oligomeric g... 29.6 2.2
cel:E01G4.6 hypothetical protein 29.3 3.2
dre:100150110 polyprotein-like 29.3 3.4
ath:AT2G39580 hypothetical protein 29.3 3.5
tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-m... 28.9 4.4
hsa:374308 PTCHD3, FLJ44037, MGC129888, PTR; patched domain co... 28.9 4.5
cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196 ... 28.9 4.7
cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooke... 28.5 6.0
mmu:65020 Zfp110, 2900024E01Rik, N28112, NRIF, Nrif1; zinc fin... 28.5 6.1
ath:AT1G61095 hypothetical protein 28.1 6.1
hsa:168455 FLJ11436, FLJ25556, MGC16181; hypothetical protein ... 28.1 6.7
ath:AT3G13210 crooked neck protein, putative / cell cycle prot... 28.1 7.0
xla:446406 ttc37, MGC83808; tetratricopeptide repeat domain 37... 27.7 8.1
ath:AT4G28200 hypothetical protein; K14557 U3 small nucleolar ... 27.7 9.2
> tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putative
; K12855 pre-mRNA-processing factor 6
Length=985
Score = 212 bits (540), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 104/122 (85%), Positives = 114/122 (93%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
ARELI+ GCQ+CPKSEDVWLEAARLEKP NAKAVLAKAVSVLPHSVRLW DAYAREKD++
Sbjct 394 ARELIATGCQQCPKSEDVWLEAARLEKPANAKAVLAKAVSVLPHSVRLWFDAYAREKDLD 453
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
R+ VLRKALEFIPNSVRLWKEA SLE+E+NARI+LTRAVECV QS E+WLALARLS+YE
Sbjct 454 QRKRVLRKALEFIPNSVRLWKEAVSLEEEKNARIMLTRAVECVPQSVEIWLALARLSSYE 513
Query 121 EA 122
EA
Sbjct 514 EA 515
Score = 34.3 bits (77), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 37/139 (26%), Positives = 66/139 (47%), Gaps = 23/139 (16%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVL-----PHSVRLWLD 51
A+++++ ++CP S ++W+ A +LE+ + ++A+A L + +WL
Sbjct 515 AQKVLNEARKKCPTSPEIWVAACKLEETQGNLKMVDTIIARARDNLIARGVAQTRDVWLR 574
Query 52 AYAREKDINDR--------RLVLRKALEFIPNSVRLWKE----ACSLEDERNARILLTRA 99
A E + + R ++ +E + N+ R+WKE A S AR L T A
Sbjct 575 -LAEEAEASGFMATCQAIVRATMKVGVEGM-NAKRIWKEDAEEALSRGSVATARALYTCA 632
Query 100 VECVSQSHEMWLALARLST 118
+E + +WLALA L T
Sbjct 633 IERLKTKKSLWLALADLET 651
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 53/126 (42%), Gaps = 19/126 (15%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLE----KPRNAKAVLAKAVSVLPHSVRLWLDA-YAR 55
AR ++ R PK+ D+W A R+E + A+ V +KAV P+S +W +A +
Sbjct 831 ARAILEKAKLRNPKNPDLWHAAIRIEVEAGNKQMAQHVASKAVQECPNSGLVWAEAIFLE 890
Query 56 EKDINDRRLVLRKALEFIPNSVRL--------WKEACSLEDERNARILLTRAVECVSQSH 107
EK + V AL N V L WKE AR L R+V +
Sbjct 891 EKSAQTHKAV--DALTKCENDVHLVLAVACLFWKEG----KISKARKWLNRSVTLDASFG 944
Query 108 EMWLAL 113
+ W A
Sbjct 945 DAWAAF 950
> tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=1032
Score = 171 bits (432), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 83/122 (68%), Positives = 100/122 (81%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
ARE+I+ GC+ CP EDVWLEAARLEKP AK++LAKA+ ++P SV+LWL+A +E +
Sbjct 333 AREIIAQGCENCPDKEDVWLEAARLEKPEYAKSILAKAIKIIPTSVKLWLEAADKETSND 392
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
+RR VLRKALEFIPNS+RLWKEA SLE+E NA ILL RAVECV +S +MWLALARL YE
Sbjct 393 NRRRVLRKALEFIPNSIRLWKEAISLENETNAYILLKRAVECVPESLDMWLALARLCPYE 452
Query 121 EA 122
EA
Sbjct 453 EA 454
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 34/64 (53%), Gaps = 0/64 (0%)
Query 12 CPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALE 71
P+S D+WL ARL A+ VL +A LP +V +W+ A E+ N+ +V R +
Sbjct 435 VPESLDMWLALARLCPYEEAQKVLNEARKKLPTNVDIWITAAKLEESNNNYEMVERIIVR 494
Query 72 FIPN 75
I N
Sbjct 495 AIDN 498
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 10/89 (11%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKP----RNAKAVLAKAVSVLPHSVRLW-----LD 51
AR+L+ RC + VW+++ +LE+ A ++ KA+ + P+ +LW L
Sbjct 666 ARKLLEKARTRC-NTPKVWMKSVQLERQLKNYEKALELVDKALEIHPYFDKLWMISGQLK 724
Query 52 AYAREKDINDRRLVLRKALEFIPNSVRLW 80
KD+ L ++ +E P SV LW
Sbjct 725 LEKHPKDVEGATLTYKQGVETCPWSVNLW 753
Score = 31.2 bits (69), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 43/93 (46%), Gaps = 3/93 (3%)
Query 29 RNAKAVLAKAVSVLPHSVRLWLDAYAREK---DINDRRLVLRKALEFIPNSVRLWKEACS 85
+ A+ +L +S + W+ A E+ I R ++ + E P+ +W EA
Sbjct 297 QKARTLLKSLISTNQKHAQGWIAAARMEELAGKIEAAREIIAQGCENCPDKEDVWLEAAR 356
Query 86 LEDERNARILLTRAVECVSQSHEMWLALARLST 118
LE A+ +L +A++ + S ++WL A T
Sbjct 357 LEKPEYAKSILAKAIKIIPTSVKLWLEAADKET 389
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 30/141 (21%), Positives = 63/141 (44%), Gaps = 27/141 (19%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLP-----HSVRLWLD 51
A+++++ ++ P + D+W+ AA+LE+ N + ++ +A+ L H WL
Sbjct 454 AQKVLNEARKKLPTNVDIWITAAKLEESNNNYEMVERIIVRAIDNLSKKGVVHIRSNWL- 512
Query 52 AYAREKDINDRRLVLRKALEFIPNSVRLW-----KEACSLEDERN---------ARILLT 97
++ + + ++ A I N++ + +++ LED AR L
Sbjct 513 ---KQAETAEANSFVKTAQSIIKNTMTIGVDEHNRKSVWLEDGETFVEHGSYECARALYK 569
Query 98 RAVECVSQSHEMWLALARLST 118
A+E + +WLAL L +
Sbjct 570 NALEYMKTRSSLWLALVELES 590
> bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, putaitve;
K12855 pre-mRNA-processing factor 6
Length=1040
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 79/122 (64%), Positives = 101/122 (82%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
ARE+I+ C++C EDVWLEAARLEKP AKAVLAKAV ++P SV++W++A RE ++N
Sbjct 337 AREIIAQACEKCGDREDVWLEAARLEKPEYAKAVLAKAVRMVPQSVKIWVEAARRESNVN 396
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
D+R +LRKALEFIPNSVRLWK+A SLEDE +A ++L RAVECV S ++WLALARL +Y+
Sbjct 397 DKRRILRKALEFIPNSVRLWKDAISLEDETDAYVMLKRAVECVPDSVDLWLALARLCSYQ 456
Query 121 EA 122
EA
Sbjct 457 EA 458
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 45/90 (50%), Gaps = 3/90 (3%)
Query 29 RNAKAVLAKAVSVLPHSVRLWLDAYAREK---DINDRRLVLRKALEFIPNSVRLWKEACS 85
+ A+ +L ++ P+ W+ A E+ I+ R ++ +A E + +W EA
Sbjct 301 KKARKLLKSVIATNPNHAPGWIAAARIEELAGKISSAREIIAQACEKCGDREDVWLEAAR 360
Query 86 LEDERNARILLTRAVECVSQSHEMWLALAR 115
LE A+ +L +AV V QS ++W+ AR
Sbjct 361 LEKPEYAKAVLAKAVRMVPQSVKIWVEAAR 390
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 13 PKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLV 65
P S D+WL ARL + A+ VL +A LP + +W+ A E+ ++++V
Sbjct 440 PDSVDLWLALARLCSYQEAQKVLNEARKHLPTNADIWITAAKLEESNGNQQMV 492
> pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855
pre-mRNA-processing factor 6
Length=1329
Score = 157 bits (398), Expect = 5e-39, Method: Composition-based stats.
Identities = 78/123 (63%), Positives = 97/123 (78%), Gaps = 1/123 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLE-KPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDI 59
A+E+I GC C K+ED+WLEA RLE K K +LAKA+ +P SV+LWL+AY +EK++
Sbjct 443 AKEIIMKGCVVCSKNEDIWLEAVRLEEKLSEVKIILAKAIKHIPTSVKLWLEAYKKEKNV 502
Query 60 NDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTY 119
+D+R VLRKA+E IPNSV+LWKEA SLE+E NA ILL RAVEC+ QS EMW+ALARL TY
Sbjct 503 DDKRKVLRKAIECIPNSVKLWKEAISLENENNAYILLKRAVECIPQSIEMWIALARLCTY 562
Query 120 EEA 122
EA
Sbjct 563 TEA 565
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 41/139 (29%), Positives = 65/139 (46%), Gaps = 20/139 (14%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLE----KPRNAKAVLAKAVSVLPHSVRLWLDAYARE 56
AR L + + + +WL A LE K + VL +AV PHS LWL A++
Sbjct 675 ARTLYNEALKIFKTKKSLWLALANLELTHGKREDVDEVLHRAVQSCPHSSVLWL-MLAKQ 733
Query 57 K----DINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRA-VECVSQSH 107
K +I+ R +L ++ N+ + A LE E N AR LL ++ V+C +
Sbjct 734 KWLNNEIDKAREILAESFIHNQNTEEISLAAIKLERENNEFDRARFLLKKSRVQC--NTP 791
Query 108 EMWLALARLS----TYEEA 122
++W+ +L Y+EA
Sbjct 792 KIWMQSVQLERLLRNYKEA 810
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 13 PKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLV 65
P+S ++W+ ARL A+ VL +A +P S +W++A E+ + ++V
Sbjct 547 PQSIEMWIALARLCTYTEAQKVLNEARKKIPTSAEIWINASQLEEKQGNIKMV 599
> mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655,
MGC36967, MGC38351, U5-102K; PRP6 pre-mRNA splicing factor
6 homolog (yeast); K12855 pre-mRNA-processing factor 6
Length=941
Score = 142 bits (357), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 75/122 (61%), Positives = 90/122 (73%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR LI G + CPKSEDVWLEAARL+ AKAV+A+AV LP SVR+++ A E DI
Sbjct 328 ARNLIMKGTEMCPKSEDVWLEAARLQPGDTAKAVVAQAVRHLPQSVRIYIRAAELETDIR 387
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
++ VLRKALE +PNSVRLWK A LE+ +ARI+L+RAVEC S E+WLALARL TYE
Sbjct 388 AKKRVLRKALEHVPNSVRLWKAAVELEEPEDARIMLSRAVECCPTSVELWLALARLETYE 447
Query 121 EA 122
A
Sbjct 448 NA 449
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 49/92 (53%), Gaps = 1/92 (1%)
Query 10 QRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKA 69
+ P S +W A LE+P +A+ +L++AV P SV LWL A AR + + R VL KA
Sbjct 398 EHVPNSVRLWKAAVELEEPEDARIMLSRAVECCPTSVELWL-ALARLETYENARKVLNKA 456
Query 70 LEFIPNSVRLWKEACSLEDERNARILLTRAVE 101
E IP +W A LE+ ++ + ++
Sbjct 457 RENIPTDRHIWITAAKLEEANGNTQMVEKIID 488
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 60/123 (48%), Gaps = 7/123 (5%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKP----RNAKAVLAKAVSVLPHSVRLWLDAY--- 53
ARE + G ++CP S +WL +RLE+ A+A+L K+ P + LWL++
Sbjct 728 AREAYNQGLKKCPHSTPLWLLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVRLE 787
Query 54 AREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLAL 113
R N ++ KAL+ PNS LW EA LE + A++ + LA+
Sbjct 788 YRAGLKNIANTLMAKALQECPNSGILWSEAVFLEARPQRKTKSVDALKKCEHDPHVLLAV 847
Query 114 ARL 116
A+L
Sbjct 848 AKL 850
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 53/111 (47%), Gaps = 13/111 (11%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAK----AVLAKAVSVLPHSVRLWLDAYARE 56
AR + + Q P + VWL AA EK + A+L +AV+ P + LWL A+
Sbjct 559 ARAIYAYALQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMG-AKS 617
Query 57 K----DINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRA 99
K D+ R +L A + PNS +W A LE E N AR LL +A
Sbjct 618 KWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKA 668
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 64/127 (50%), Gaps = 12/127 (9%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR +++L Q P SE++WL A +LE N A+ +LAKA S P + R+++ + E
Sbjct 627 ARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKARSSAP-TARVFMKSVKLE 685
Query 57 ---KDINDRRLVLRKALEFIPNSVRLWKEACSLEDE----RNARILLTRAVECVSQSHEM 109
+I+ + + +AL + +LW +E++ AR + ++ S +
Sbjct 686 WVLGNISAAQELCEEALRHYEDFPKLWMMKGQIEEQGELMEKAREAYNQGLKKCPHSTPL 745
Query 110 WLALARL 116
WL L+RL
Sbjct 746 WLLLSRL 752
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR ++S + CP S ++WL ARLE NA+ VL KA +P +W+ A E+
Sbjct 419 ARIMLSRAVECCPTSVELWLALARLETYENARKVLNKARENIPTDRHIWITAAKLEEANG 478
Query 61 DRRLV 65
+ ++V
Sbjct 479 NTQMV 483
> hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6;
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae);
K12855 pre-mRNA-processing factor 6
Length=941
Score = 142 bits (357), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 75/122 (61%), Positives = 90/122 (73%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR LI G + CPKSEDVWLEAARL+ AKAV+A+AV LP SVR+++ A E DI
Sbjct 328 ARNLIMKGTEMCPKSEDVWLEAARLQPGDTAKAVVAQAVRHLPQSVRIYIRAAELETDIR 387
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
++ VLRKALE +PNSVRLWK A LE+ +ARI+L+RAVEC S E+WLALARL TYE
Sbjct 388 AKKRVLRKALEHVPNSVRLWKAAVELEEPEDARIMLSRAVECCPTSVELWLALARLETYE 447
Query 121 EA 122
A
Sbjct 448 NA 449
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 49/92 (53%), Gaps = 1/92 (1%)
Query 10 QRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKA 69
+ P S +W A LE+P +A+ +L++AV P SV LWL A AR + + R VL KA
Sbjct 398 EHVPNSVRLWKAAVELEEPEDARIMLSRAVECCPTSVELWL-ALARLETYENARKVLNKA 456
Query 70 LEFIPNSVRLWKEACSLEDERNARILLTRAVE 101
E IP +W A LE+ ++ + ++
Sbjct 457 RENIPTDRHIWITAAKLEEANGNTQMVEKIID 488
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 63/125 (50%), Gaps = 11/125 (8%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKP----RNAKAVLAKAVSVLPHSVRLWLDAYARE 56
ARE + G ++CP S +WL +RLE+ A+A+L K+ P + LWL++ E
Sbjct 728 AREAYNQGLKKCPHSTPLWLLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVRLE 787
Query 57 -----KDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWL 111
K+I + ++ KAL+ PNS LW EA LE R A++ + L
Sbjct 788 YRAGLKNIAN--TLMAKALQECPNSGILWSEAIFLEARPQRRTKSVDALKKCEHDPHVLL 845
Query 112 ALARL 116
A+A+L
Sbjct 846 AVAKL 850
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 53/111 (47%), Gaps = 13/111 (11%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAK----AVLAKAVSVLPHSVRLWLDAYARE 56
AR + + Q P + VWL AA EK + A+L +AV+ P + LWL A+
Sbjct 559 ARAIYAYALQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMG-AKS 617
Query 57 K----DINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRA 99
K D+ R +L A + PNS +W A LE E + AR LL +A
Sbjct 618 KWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKA 668
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/127 (27%), Positives = 64/127 (50%), Gaps = 12/127 (9%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR +++L Q P SE++WL A +LE + A+ +LAKA S P + R+++ + E
Sbjct 627 ARSILALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKARSSAP-TARVFMKSVKLE 685
Query 57 ---KDINDRRLVLRKALEFIPNSVRLWKEACSLEDER----NARILLTRAVECVSQSHEM 109
+I + + +AL + +LW +E+++ AR + ++ S +
Sbjct 686 WVQDNIRAAQDLCEEALRHYEDFPKLWMMKGQIEEQKEMMEKAREAYNQGLKKCPHSTPL 745
Query 110 WLALARL 116
WL L+RL
Sbjct 746 WLLLSRL 752
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR ++S + CP S ++WL ARLE NA+ VL KA +P +W+ A E+
Sbjct 419 ARIMLSRAVECCPTSVELWLALARLETYENARKVLNKARENIPTDRHIWITAAKLEEANG 478
Query 61 DRRLV 65
+ ++V
Sbjct 479 NTQMV 483
> xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor
6 homolog; K12855 pre-mRNA-processing factor 6
Length=948
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 73/122 (59%), Positives = 90/122 (73%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR LI G + CPKSEDVWLEAARL+ AKAV+A+AV LP SVR+++ A E D+
Sbjct 335 ARNLIMKGTEMCPKSEDVWLEAARLQPGDTAKAVVAQAVRHLPQSVRIYIRAAELETDLR 394
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
++ VLRKALE +PNSVRLWK A LE+ +ARI+L+RAVEC + E+WLALARL TYE
Sbjct 395 AKKRVLRKALEHVPNSVRLWKAAVELEEPEDARIMLSRAVECCPTNVELWLALARLETYE 454
Query 121 EA 122
A
Sbjct 455 NA 456
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 49/92 (53%), Gaps = 1/92 (1%)
Query 10 QRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKA 69
+ P S +W A LE+P +A+ +L++AV P +V LWL A AR + + R VL KA
Sbjct 405 EHVPNSVRLWKAAVELEEPEDARIMLSRAVECCPTNVELWL-ALARLETYENARKVLNKA 463
Query 70 LEFIPNSVRLWKEACSLEDERNARILLTRAVE 101
E IP +W A LE+ ++ + ++
Sbjct 464 RENIPTDRHIWITAAKLEEANGNTQMVEKIID 495
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 53/111 (47%), Gaps = 13/111 (11%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAK----AVLAKAVSVLPHSVRLWLDAYARE 56
AR + + Q P + VWL AA EK + A+L +AV+ P + LWL A+
Sbjct 566 ARAIYAHSLQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMG-AKS 624
Query 57 K----DINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRA 99
K D+ R +L A + PNS +W A LE E N AR LL +A
Sbjct 625 KWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKA 675
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 38/125 (30%), Positives = 62/125 (49%), Gaps = 11/125 (8%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR+ + G ++C S +WL +RLE+ A+A+L K+ P + LWL++ E
Sbjct 735 ARDAYNQGLKKCIHSTSLWLLLSRLEEKVGQLTRARAILEKSRLKNPKTPELWLESVRLE 794
Query 57 -----KDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWL 111
K+I + ++ KAL+ PNS LW EA LE + A++ + L
Sbjct 795 FRAGLKNIAN--TLMAKALQECPNSGILWAEAVFLEARPQRKTKSVDALKKCEHDPHVLL 852
Query 112 ALARL 116
A+A+L
Sbjct 853 AVAKL 857
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 63/127 (49%), Gaps = 12/127 (9%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR +++L Q P SE++WL A +LE N A+ +LAKA S P + R+++ + E
Sbjct 634 ARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKARSSAP-TARVFMKSVKLE 692
Query 57 ---KDINDRRLVLRKALEFIPNSVRLWKEACSLEDE----RNARILLTRAVECVSQSHEM 109
+I + + +AL + +LW +E++ AR + ++ S +
Sbjct 693 WVLGNIEAAQDLCEEALRHYEDFPKLWMMKGQIEEQMEQTEKARDAYNQGLKKCIHSTSL 752
Query 110 WLALARL 116
WL L+RL
Sbjct 753 WLLLSRL 759
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR ++S + CP + ++WL ARLE NA+ VL KA +P +W+ A E+
Sbjct 426 ARIMLSRAVECCPTNVELWLALARLETYENARKVLNKARENIPTDRHIWITAAKLEEANG 485
Query 61 DRRLV 65
+ ++V
Sbjct 486 NTQMV 490
> ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing
factor, transesterification mechanism; K12855 pre-mRNA-processing
factor 6
Length=1029
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 69/122 (56%), Positives = 87/122 (71%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR I GC+ CPK+EDVWLEA RL P +AK V+AK V ++P+SV+LWL+A E D+
Sbjct 406 ARFQIQRGCEECPKNEDVWLEACRLANPEDAKGVIAKGVKLIPNSVKLWLEAAKLEHDVE 465
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
++ VLRK LE IP+SVRLWK L +E +ARILL RAVEC E+W+ALARL TY
Sbjct 466 NKSRVLRKGLEHIPDSVRLWKAVVELANEEDARILLHRAVECCPLHLELWVALARLETYA 525
Query 121 EA 122
E+
Sbjct 526 ES 527
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 38/96 (39%), Positives = 55/96 (57%), Gaps = 13/96 (13%)
Query 18 VWLEAARLEKPRNAK----AVLAKAVSVLPHSVRLWLDAYAREK----DINDRRLVLRKA 69
+WL+AA+LEK ++ A+L KAV+ +P + LWL A+EK D+ R +L++A
Sbjct 661 IWLKAAQLEKSHGSRESLDALLRKAVTYVPQAEVLWLMG-AKEKWLAGDVPAARAILQEA 719
Query 70 LEFIPNSVRLWKEACSLEDERN----ARILLTRAVE 101
IPNS +W A LE E AR+LL +A E
Sbjct 720 YAAIPNSEEIWLAAFKLEFENKEPERARMLLAKARE 755
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 59/128 (46%), Gaps = 14/128 (10%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLE----KPRNAKAVLAKAVSVLPHSVRLWLDAYARE 56
AR ++ P SE++WL A +LE +P A+ +LAKA + R+W+ + E
Sbjct 712 ARAILQEAYAAIPNSEEIWLAAFKLEFENKEPERARMLLAKARE-RGGTERVWMKSAIVE 770
Query 57 KDIND---RRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSH-----E 108
+++ + R +L + L+ P +LW LE ER + R H
Sbjct 771 RELGNVEEERRLLNEGLKQFPTFFKLWLMLGQLE-ERFKHLEQARKAYDTGLKHCPHCIP 829
Query 109 MWLALARL 116
+WL+LA L
Sbjct 830 LWLSLADL 837
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 41/87 (47%), Gaps = 7/87 (8%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR+ G + CP +WL A LE+ N A+A+L A P LWL A E
Sbjct 813 ARKAYDTGLKHCPHCIPLWLSLADLEEKVNGLNKARAILTTARKKNPGGAELWLAAIRAE 872
Query 57 KDINDRR---LVLRKALEFIPNSVRLW 80
+++R ++ KAL+ P S LW
Sbjct 873 LRHDNKREAEHLMSKALQDCPKSGILW 899
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 43/78 (55%), Gaps = 5/78 (6%)
Query 43 PHSVRLWLDAYAREKDINDR----RLVLRKALEFIPNSVRLWKEACSLEDERNARILLTR 98
P + W+ A AR ++++ + R +++ E P + +W EAC L + +A+ ++ +
Sbjct 384 PKNPNGWI-AAARVEEVDGKIKAARFQIQRGCEECPKNEDVWLEACRLANPEDAKGVIAK 442
Query 99 AVECVSQSHEMWLALARL 116
V+ + S ++WL A+L
Sbjct 443 GVKLIPNSVKLWLEAAKL 460
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDA 52
AR L+ + CP ++W+ ARLE +K VL KA LP +W+ A
Sbjct 497 ARILLHRAVECCPLHLELWVALARLETYAESKKVLNKAREKLPKEPAIWITA 548
> cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=968
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 71/122 (58%), Positives = 86/122 (70%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR I GC++ SE++WL A RL P ++++A AV PHSVRLW A E+D+
Sbjct 354 ARNFIMEGCEKIKNSEELWLHAIRLHPPELGRSIVANAVRSCPHSVRLWCKASDLEQDLK 413
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
D++ VLRKALE IP+SV+LWK A LED +ARILLTRAVEC S S EMWLALARL TYE
Sbjct 414 DKKKVLRKALEQIPSSVKLWKAAVELEDPEDARILLTRAVECCSSSTEMWLALARLETYE 473
Query 121 EA 122
A
Sbjct 474 NA 475
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 47/91 (51%), Gaps = 1/91 (1%)
Query 10 QRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKA 69
++ P S +W A LE P +A+ +L +AV S +WL A AR + + R VL KA
Sbjct 424 EQIPSSVKLWKAAVELEDPEDARILLTRAVECCSSSTEMWL-ALARLETYENARKVLNKA 482
Query 70 LEFIPNSVRLWKEACSLEDERNARILLTRAV 100
E IP +W A LE+ R + ++ + V
Sbjct 483 REHIPTDRHIWLSAARLEETRGQKDMVDKIV 513
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR L++ + C S ++WL ARLE NA+ VL KA +P +WL A E+
Sbjct 445 ARILLTRAVECCSSSTEMWLALARLETYENARKVLNKAREHIPTDRHIWLSAARLEETRG 504
Query 61 DRRLV 65
+ +V
Sbjct 505 QKDMV 509
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 28/46 (60%), Gaps = 4/46 (8%)
Query 11 RCPKSEDVWLEAARLEK----PRNAKAVLAKAVSVLPHSVRLWLDA 52
R PK++D+WLE+ R E+ P AK +++A+ S +LW +A
Sbjct 799 RNPKNDDLWLESVRFEQRVGCPEMAKERMSRALQECEGSGKLWAEA 844
Score = 31.6 bits (70), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 32/123 (26%), Positives = 55/123 (44%), Gaps = 7/123 (5%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR + G ++CP +W+ RLE+ A+ L KA P + LWL++ E
Sbjct 755 ARLAYTQGIRKCPGVIPLWILLVRLEEKAGQIVKARVDLEKARLRNPKNDDLWLESVRFE 814
Query 57 KDINDRRLV---LRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLAL 113
+ + + + +AL+ S +LW EA +E R A++ + + +A
Sbjct 815 QRVGCPEMAKERMSRALQECEGSGKLWAEAIWMEGPHGRRAKSIDALKKCEHNPHVLIAA 874
Query 114 ARL 116
ARL
Sbjct 875 ARL 877
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 4/46 (8%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAK----AVLAKAVSVL 42
AR++++ + P +WL AARLE+ R K ++AKA+S L
Sbjct 475 ARKVLNKAREHIPTDRHIWLSAARLEETRGQKDMVDKIVAKAMSSL 520
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 37/95 (38%), Gaps = 11/95 (11%)
Query 30 NAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDE 89
+ K L S++P +DI R++L+ E P W + LE++
Sbjct 295 DPKGYLTDMQSIIPQ-------MGGDLQDIKKARMLLKSVRETNPRHPPAWVASAVLEEQ 347
Query 90 ----RNARILLTRAVECVSQSHEMWLALARLSTYE 120
+ AR + E + S E+WL RL E
Sbjct 348 AGKLQTARNFIMEGCEKIKNSEELWLHAIRLHPPE 382
> dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02,
zgc:65913; c20orf14 homolog (H. sapiens); K12855 pre-mRNA-processing
factor 6
Length=944
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 73/122 (59%), Positives = 88/122 (72%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR LI G + CPKSEDVWLEAARL+ AKAV+A+AV LP SVR+++ A E DI
Sbjct 331 ARNLIMKGTEMCPKSEDVWLEAARLQPGDTAKAVVAQAVRHLPQSVRIYIRAAELETDIR 390
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
++ VLRKALE + SVRLWK A LE+ +ARI+L+RAVEC S E+WLALARL TYE
Sbjct 391 AKKRVLRKALENVSKSVRLWKTAVELEEPEDARIMLSRAVECCPTSVELWLALARLETYE 450
Query 121 EA 122
A
Sbjct 451 NA 452
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 48/88 (54%), Gaps = 1/88 (1%)
Query 14 KSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFI 73
KS +W A LE+P +A+ +L++AV P SV LWL A AR + + R VL KA E I
Sbjct 405 KSVRLWKTAVELEEPEDARIMLSRAVECCPTSVELWL-ALARLETYENARRVLNKARENI 463
Query 74 PNSVRLWKEACSLEDERNARILLTRAVE 101
P +W A LE+ ++ + ++
Sbjct 464 PTDRHIWITAAKLEEANGNTQMVEKIID 491
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 41/123 (33%), Positives = 60/123 (48%), Gaps = 7/123 (5%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAY--- 53
ARE + G ++CP S +WL +RLE+ A+A+L KA P S LWL++
Sbjct 731 AREAYNQGLKKCPHSMSLWLLLSRLEEKVGQLTRARAILEKARLKNPQSPELWLESVRLE 790
Query 54 AREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLAL 113
R N ++ KAL+ PNS LW EA LE + A++ + LA+
Sbjct 791 YRAGLKNIANTLMAKALQECPNSGILWSEAVFLEARPQRKTKSVDALKKCEHDPHVLLAV 850
Query 114 ARL 116
A+L
Sbjct 851 AKL 853
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 53/111 (47%), Gaps = 13/111 (11%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAK----AVLAKAVSVLPHSVRLWLDAYARE 56
AR + + Q P + VWL AA EK + A+L +AV+ P + LWL A+
Sbjct 562 ARAIYAHALQVFPSKKSVWLRAAYFEKNNGTRESLEALLQRAVAHCPKAEVLWLMG-AKS 620
Query 57 K----DINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRA 99
K D+ R +L A + PNS +W A LE E N AR LL +A
Sbjct 621 KWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKA 671
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 63/127 (49%), Gaps = 12/127 (9%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR +++L Q P SE++WL A +LE N A+ +LAKA S P + R+++ + E
Sbjct 630 ARSILALAFQANPNSEEIWLAAVKLESENNEYERARRLLAKARSSAP-TARVFMKSVRLE 688
Query 57 ---KDINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRAVECVSQSHEM 109
+I + +AL+ + +LW +E++ AR + ++ S +
Sbjct 689 WVLGNIEAAHELCTEALKHYEDFPKLWMMRGQIEEQSESIDRAREAYNQGLKKCPHSMSL 748
Query 110 WLALARL 116
WL L+RL
Sbjct 749 WLLLSRL 755
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
AR ++S + CP S ++WL ARLE NA+ VL KA +P +W+ A E+
Sbjct 422 ARIMLSRAVECCPTSVELWLALARLETYENARRVLNKARENIPTDRHIWITAAKLEEANG 481
Query 61 DRRLV 65
+ ++V
Sbjct 482 NTQMV 486
> cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat
protein ; K12855 pre-mRNA-processing factor 6
Length=923
Score = 125 bits (315), Expect = 2e-29, Method: Composition-based stats.
Identities = 62/122 (50%), Positives = 90/122 (73%), Gaps = 0/122 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDIN 60
ARE+I+ GC+ CPK+ED+WLEA RL KP ++ K++ +P+S ++W+ A RE + N
Sbjct 291 AREIIAKGCEMCPKNEDIWLEAIRLGKPEQIDKIIVKSIKFIPNSTKVWMVAANRETNKN 350
Query 61 DRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLALARLSTYE 120
+ L+++KALEFIPNS++LWKEA SL D + + LL++AV+CV QS E+WL ARLS Y
Sbjct 351 KKLLIIKKALEFIPNSIKLWKEAISLVDNESEKALLSKAVKCVPQSEELWLRYARLSEYC 410
Query 121 EA 122
+A
Sbjct 411 DA 412
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/82 (37%), Positives = 46/82 (56%), Gaps = 2/82 (2%)
Query 13 PKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEF 72
P S +W EA L + KA+L+KAV +P S LWL YAR + D + +L +A +
Sbjct 364 PNSIKLWKEAISLVDNESEKALLSKAVKCVPQSEELWL-RYARLSEYCDAQKILNEARKV 422
Query 73 IPNSVRLWKEACSLEDERNARI 94
+P +W EA LE E+N ++
Sbjct 423 LPTFPGIWVEAAKLE-EQNGKV 443
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 2 RELISLGCQRCPKSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREK 57
+ L+S + P+SE++WL ARL + +A+ +L +A VLP +W++A E+
Sbjct 383 KALLSKAVKCVPQSEELWLRYARLSEYCDAQKILNEARKVLPTFPGIWVEAAKLEE 438
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/97 (20%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREK---DINDRRLVLRKALEFIPNSVRLWK 81
L + A+ +L V+ P W+ A E+ ++ R ++ K E P + +W
Sbjct 251 LSDIKKARLLLKSVVNTNPKHSPGWIAAARFEEFVGRLSHAREIIAKGCEMCPKNEDIWL 310
Query 82 EACSLEDERNARILLTRAVECVSQSHEMWLALARLST 118
EA L ++ ++++ + S ++W+ A T
Sbjct 311 EAIRLGKPEQIDKIIVKSIKFIPNSTKVWMVAANRET 347
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 27/115 (23%), Positives = 47/115 (40%), Gaps = 13/115 (11%)
Query 15 SEDVWLEAARLEKPRNAKAVL----AKAVSVLPHSVRLWL-----DAYAREKDINDRRLV 65
S +W+E+ +LE + + +++V P S LWL A IN+ +
Sbjct 638 SVQIWVESIKLENDQKNYDLCILYCSESVKEYPSSPNLWLLYGFIYRKAFPDRINEALKI 697
Query 66 LRKALEFIPNSVRLWKEACS----LEDERNARILLTRAVECVSQSHEMWLALARL 116
+ L F +S+ LW L++ + AR L A E+W+ +L
Sbjct 698 YEEGLNFCSDSIELWFSTIELLMLLQNWKKARTFLDLARSKNKNQPELWMQTIKL 752
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 29/118 (24%), Positives = 56/118 (47%), Gaps = 21/118 (17%)
Query 10 QRCPKSEDVWLEAARLEKPRN----AKAVLAKAVS--------VLPHSVRLWLDAYAREK 57
+ CP + +WL+AA+ + A+ +L+K S ++ + RL L +
Sbjct 565 KNCPDKQILWLKAAQNQSANGNAEIARLILSKGYSSSLNDKEEIVLEAARLELS----QG 620
Query 58 DINDRRLVLRKALEFIPNSVRLWKEACSLE-DERNARILLTRAVECVSQ---SHEMWL 111
+I +++L + P SV++W E+ LE D++N + + E V + S +WL
Sbjct 621 EIERAKIILERERTNSP-SVQIWVESIKLENDQKNYDLCILYCSESVKEYPSSPNLWL 677
> sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-processing
factor 6
Length=899
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 68/125 (54%), Gaps = 3/125 (2%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKP--RNAKAVLAKAVSVLPHSVRLWLDAYAREKD 58
A+++I GCQ CP+S D+WLE RL + K ++A A++ P S LW A E
Sbjct 260 AKKIIENGCQECPRSSDIWLENIRLHESDVHYCKTLVATAINFNPTSPLLWFKAIDLEST 319
Query 59 INDRRLVLRKALEFIPNSVRLWKEACSLEDERNARI-LLTRAVECVSQSHEMWLALARLS 117
++ V+RKAL+ IP LWK A S E ++ I +L +A + + QS ++ A L
Sbjct 320 TVNKYRVVRKALQEIPRDEGLWKLAVSFEADKAQVIKMLEKATQFIPQSMDLLTAYTNLQ 379
Query 118 TYEEA 122
+Y A
Sbjct 380 SYHNA 384
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 43/98 (43%), Gaps = 2/98 (2%)
Query 10 QRCPKSEDVWLEAARLEKPR-NAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRK 68
Q P+ E +W A E + +L KA +P S+ L L AY + ++ ++ L
Sbjct 332 QEIPRDEGLWKLAVSFEADKAQVIKMLEKATQFIPQSMDL-LTAYTNLQSYHNAKMTLNS 390
Query 69 ALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQS 106
+ +P +W + LE+ N I + + V + +
Sbjct 391 FRKILPQEPEIWIISTLLEERNNPDIPVDKLVSLLKEG 428
> hsa:653889 pre-mRNA-processing factor 6-like
Length=406
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/123 (32%), Positives = 60/123 (48%), Gaps = 7/123 (5%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKP----RNAKAVLAKAVSVLPHSVRLWLDAY--- 53
ARE + G ++CP S +WL +RLE+ A+A+L K+ P + LWL++
Sbjct 193 AREAYNQGLKKCPHSTPLWLLLSRLEEKIGQLTRARAILEKSRLKNPKNPGLWLESVRLE 252
Query 54 AREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLAL 113
R N ++ KAL+ PNS LW EA LE R A++ + LA+
Sbjct 253 YRAGLKNIANTLMAKALQECPNSGILWSEAIFLEARPQRRTKSVDALKKCEHDPHVLLAV 312
Query 114 ARL 116
A+L
Sbjct 313 AKL 315
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 53/111 (47%), Gaps = 13/111 (11%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRNAK----AVLAKAVSVLPHSVRLWLDAYARE 56
AR + + Q P + VWL AA EK + A+L +AV+ P + LWL A+
Sbjct 24 ARAIYAYALQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMG-AKS 82
Query 57 K----DINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRA 99
K D+ R +L A + PNS +W A LE E + AR LL +A
Sbjct 83 KWLAGDVPAARSILALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKA 133
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/127 (27%), Positives = 64/127 (50%), Gaps = 12/127 (9%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
AR +++L Q P SE++WL A +LE + A+ +LAKA S P + R+++ + E
Sbjct 92 ARSILALAFQANPNSEEIWLAAVKLESENDEYERARRLLAKARSSAP-TARVFMKSVKLE 150
Query 57 ---KDINDRRLVLRKALEFIPNSVRLWKEACSLEDER----NARILLTRAVECVSQSHEM 109
+I + + +AL + +LW +E+++ AR + ++ S +
Sbjct 151 WVQDNIRAAQDLCEEALRHYEDFPKLWMMKGQIEEQKEMMEKAREAYNQGLKKCPHSTPL 210
Query 110 WLALARL 116
WL L+RL
Sbjct 211 WLLLSRL 217
> dre:100149473 polyprotein-like
Length=2049
Score = 35.0 bits (79), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFI 73
L++P + V+ K V VL H+ R L+AYA D ++R ++L A +++
Sbjct 749 LDRPNRPQQVMLKVVKVLIHNARHSLEAYALLDDGSERTILLSSATQYL 797
> dre:100151459 Gag-Pol polyprotein-like
Length=2050
Score = 35.0 bits (79), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFI 73
L++P + V+ K V VL H+ R L+AYA D ++R ++L A +++
Sbjct 810 LDRPNRPQQVMLKVVKVLIHNARHSLEAYALLDDGSERTILLSSATQYL 858
> dre:100333340 polyprotein-like
Length=1609
Score = 35.0 bits (79), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFI 73
L++P + V+ K V VL H+ R L+AYA D ++R ++L A +++
Sbjct 274 LDRPNRPQQVMLKVVKVLIHNARHSLEAYALLDDGSERTILLSSATQYL 322
> dre:100331348 polyprotein-like
Length=1574
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFI 73
L++P + V+ K V VL H+ R L+AYA D ++R ++L A +++
Sbjct 274 LDRPNRPQQVMLKVVKVLLHNARHSLEAYALLDDGSERTILLSSATQYL 322
> cel:F25B4.5 hypothetical protein; K13217 pre-mRNA-processing
factor 39
Length=710
Score = 34.3 bits (77), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 41/101 (40%), Gaps = 16/101 (15%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLEKPRN----AKAVLAKAVSVLPHSVRLWLDAYARE 56
ARE R P W + A EK AKAV K + +P S+ LWL A
Sbjct 118 AREKYRSFLSRYPNCYGFWQKYAEYEKKMGNIAEAKAVWEKGIISIPLSIDLWLGYTADV 177
Query 57 KDIND------RRLVLR----KALEFIPNSVRLWKEACSLE 87
K+I + R L R LE+ S RLW EA E
Sbjct 178 KNIKNFPPESLRDLYARAIEIAGLEY--QSDRLWLEAIGFE 216
> sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck
Length=687
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 42/78 (53%), Gaps = 8/78 (10%)
Query 18 VWLEAARL-----EKPRNAKAVLAKAVSVLPH--SVRLWLDAYAREKDINDRRLVLRKAL 70
+WL A+ + P+ A+ +L KA+ + P + + +++ + K+ + R + K +
Sbjct 406 IWLMYAKFLIRHDDVPK-ARKILGKAIGLCPKAKTFKGYIELEVKLKEFDRVRKIYEKFI 464
Query 71 EFIPNSVRLWKEACSLED 88
EF P+ +++W + LE+
Sbjct 465 EFQPSDLQIWSQYGELEE 482
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 26/102 (25%), Positives = 47/102 (46%), Gaps = 14/102 (13%)
Query 19 WLEAARLE----KPRNAKAVLAKAVSVLPHSVRLWL---DAYAREKDINDRRLVLRKALE 71
W+ A+ E R A+++ +A+ V + LW+ DA + K IN R ++ +A+
Sbjct 68 WIRYAQFEIEQHDMRRARSIFERALLVDSSFIPLWIRYIDAELKVKCINHARNLMNRAIS 127
Query 72 FIPNSVRLWKEACSLEDERNARILLTRAVECVSQSHEMWLAL 113
+P +LW + +E+ N VE V + W +L
Sbjct 128 TLPRVDKLWYKYLIVEESLN-------NVEIVRSLYTKWCSL 162
> mmu:170936 Zfp369, B930030B22Rik, D230020H11Rik, NRIF2; zinc
finger protein 369; K12458 zinc finger protein 274
Length=845
Score = 32.3 bits (72), Expect = 0.38, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 40/84 (47%), Gaps = 2/84 (2%)
Query 37 KAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILL 96
K+VS L WL R K LVL + L +P +R+W E+ ED + A + L
Sbjct 198 KSVSQLRKLCHQWLQPSTRSKKQILELLVLEQFLNALPEKLRVWVESQHPEDCK-AVVAL 256
Query 97 TRAVECVSQSHEMWLALARLSTYE 120
+ VS+ + WLA + +T E
Sbjct 257 LENMTSVSKD-DAWLACSSEATDE 279
> dre:100002224 polyprotein-like
Length=2022
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALE 71
L++P + V+ K V VL HS R ++ +A D ++R LVL+ ++
Sbjct 713 LDRPNRSPKVMLKVVKVLLHSGRKTMETHAVLDDGSERTLVLQPVVQ 759
> dre:796060 polyprotein-like
Length=1793
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALE 71
L++P + V+ K V VL HS R ++ +A D ++R LVL+ ++
Sbjct 484 LDRPNRSPKVMLKVVKVLLHSGRKTMETHAVLDDGSERTLVLQPVVQ 530
> bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain
containing protein; K12869 crooked neck
Length=665
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 28/67 (41%), Gaps = 4/67 (5%)
Query 54 AREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDERN----ARILLTRAVECVSQSHEM 109
A ++D R V +AL+ PN+V LW E + AR L R V + + +
Sbjct 84 ANQQDFRRARSVFERALQVDPNNVNLWLRYIETEMKNKNVNAARNLFDRVVSLLPRVDQF 143
Query 110 WLALARL 116
W A
Sbjct 144 WFKYAHF 150
> dre:100002091 polyprotein-like
Length=1966
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALE 71
L++P + V+ K V VL HS R ++ +A D ++R LVL+ ++
Sbjct 657 LDRPNRSPKVMLKVVKVLLHSGRKTIETHAVLDDGSERTLVLQPVVK 703
> cel:M03F8.3 hypothetical protein; K12869 crooked neck
Length=747
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 24/114 (21%), Positives = 56/114 (49%), Gaps = 12/114 (10%)
Query 14 KSEDVWLEAARLE----KPRNAKAVLAKAVSVLPHSVRLWLD-AYARE--KDINDRRLVL 66
+S +WL+ A +E + +A+ V +A++++P +++ WL +Y E ++I R +
Sbjct 118 RSISIWLQYAEMEMRCKQINHARNVFDRAITIMPRAMQFWLKYSYMEEVIENIPGARQIF 177
Query 67 RKALEFIPNSVRLWKEACSLE----DERNARILLTRAVECVSQSHEMWLALARL 116
+ +E+ P + W+ + E + AR + R + + + W+ A+
Sbjct 178 ERWIEWEPPE-QAWQTYINFELRYKEIDRARSVYQRFLHVHGINVQNWIKYAKF 230
> hsa:22796 COG2, LDLC; component of oligomeric golgi complex
2
Length=737
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 36/63 (57%), Gaps = 2/63 (3%)
Query 23 ARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKE 82
A ++K R+A+A++ + V V P+ + ++ + E N +++ K LEF+P+ RL +E
Sbjct 235 ATIDKTRDAEALVGQ-VLVKPYIDEVIIEQFV-ESHPNGLQVMYNKLLEFVPHHCRLLRE 292
Query 83 ACS 85
Sbjct 293 VTG 295
> cpv:cgd1_3160 mRNA 3' end processing protein RNA14, HAT repeats
Length=1452
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 30/66 (45%), Gaps = 7/66 (10%)
Query 43 PHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKEACS-LEDERNA--RILLTRA 99
P+ LW + + + V +ALEF P S +WK L+ ++N ++LL
Sbjct 17 PYDYSLWENIFT----VKSESEVFERALEFFPTSPIVWKRYIEYLQSQKNTDEKVLLGIY 72
Query 100 VECVSQ 105
C+ Q
Sbjct 73 QRCIHQ 78
> dre:406663 cog2, zC8A9.2, zgc:56436; component of oligomeric
golgi complex 2
Length=730
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 36/63 (57%), Gaps = 2/63 (3%)
Query 23 ARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKE 82
A ++K R+A+A++ + V V P+ + ++ + + N +++ K LEF+P+ RL +E
Sbjct 230 ATIDKTRDAEALVGQ-VLVKPYMDEVIVEQFVKSSP-NGLKVMYAKLLEFVPHHCRLLRE 287
Query 83 ACS 85
Sbjct 288 VTG 290
> cel:E01G4.6 hypothetical protein
Length=884
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query 45 SVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKEACSLE 87
++ L L+A KD+N R LV +ALEF+ RL KE E
Sbjct 49 AISLTLEA----KDVNIRDLVKYRALEFVGCQDRLLKEVVDFE 87
> dre:100150110 polyprotein-like
Length=1392
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 25 LEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALE 71
L++P ++ V+ K V VL H ++AYA D ++R +VL + ++
Sbjct 173 LDRPNRSQKVMLKIVKVLLHHKEKAMEAYAVLDDGSERSIVLSQVVD 219
> ath:AT2G39580 hypothetical protein
Length=1577
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query 30 NAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRL----VLRKALEFIPNSVRLWK 81
N + ++ A+ V P + W++A DIN + L+KAL P SV+LW+
Sbjct 1495 NLASTISCAIPVAPEYI--WVEAGEIVSDINGFKTRAERFLKKALSVYPMSVKLWR 1548
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 5/39 (12%)
Query 16 EDVWLEAARLEKPRN-----AKAVLAKAVSVLPHSVRLW 49
E +W+EA + N A+ L KA+SV P SV+LW
Sbjct 1509 EYIWVEAGEIVSDINGFKTRAERFLKKALSVYPMSVKLW 1547
> tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-mRNA-splicing
factor SYF1
Length=966
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 41/85 (48%), Gaps = 16/85 (18%)
Query 43 PHSVRLWLDAYAREKDI--NDRRLVLRKALEFIPNSVRLW----KEACS-------LEDE 89
P V++W+ +KD R L+ +AL +P S +LW KE + LED
Sbjct 55 PFQVKVWVGYLNSKKDAPPYTRFLLYERALRGLPGSYKLWFAYLKERVASLSSHDPLEDS 114
Query 90 R---NARILLTRAVECVSQSHEMWL 111
R A ++ RA+ +S+ ++W+
Sbjct 115 RPFEEANVVFERALVHLSRMPKIWM 139
> hsa:374308 PTCHD3, FLJ44037, MGC129888, PTR; patched domain
containing 3
Length=767
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 29/67 (43%), Gaps = 6/67 (8%)
Query 14 KSEDVWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAR-----EKDINDRRLVLRK 68
K D W + R +K N + K V V + WLDAY + +D N++ +
Sbjct 663 KKVDYWDKDVR-QKLENCTKIFEKNVYVDKNLTEFWLDAYVQYLKGNSQDPNEKNTFMNN 721
Query 69 ALEFIPN 75
+F+ N
Sbjct 722 IPDFLSN 728
> cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196
vacuolar protein-sorting-associated protein 4
Length=462
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Query 66 LRKALEFIPNSVRLWKEACSLE-DERNARILLTRAVECVSQSHEM 109
L +AL ++++ W C + DER ++LLTR + VS++ ++
Sbjct 23 LEEALNIYISALQKWDHICKYQNDERVKKVLLTRMEQLVSRAEQI 67
> cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooked
neck
Length=736
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Query 56 EKDINDRRLVLRKALEFIPNSVRLWKEACSLE----DERNARILLTRAVECVSQSHEMWL 111
+ +I + R + + + +VR+W+E LE + NAR L R + + E W+
Sbjct 115 QNNIKNSRSIFERGILVNYENVRIWREYIKLEITNGNINNARNLFERVTHLLPRIDEFWI 174
> mmu:65020 Zfp110, 2900024E01Rik, N28112, NRIF, Nrif1; zinc finger
protein 110; K12458 zinc finger protein 274
Length=832
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 26/60 (43%), Gaps = 0/60 (0%)
Query 37 KAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDERNARILL 96
KA+S L WL R K LVL + L +P R+W E+ ED + LL
Sbjct 178 KAMSQLRKLCHQWLQPNTRSKKQILELLVLEQFLNALPEKFRVWVESQHPEDCKAVVALL 237
> ath:AT1G61095 hypothetical protein
Length=125
Score = 28.1 bits (61), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 31/57 (54%), Gaps = 4/57 (7%)
Query 49 WLDAYAREKDINDRRLVLRKALEFIPNSVR-LWKEACSLEDERNARILLTRAVECVS 104
+ + Y + +N +++K ++P SVR ++++ +L DE + LT A EC S
Sbjct 39 FENCYTENETVNH---IVQKFPSYVPKSVRSFFQKSIALIDEESREAYLTDAEECAS 92
> hsa:168455 FLJ11436, FLJ25556, MGC16181; hypothetical protein
FLJ36031
Length=235
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 10/84 (11%)
Query 21 EAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRLVLRKALEFIPNSVRLW 80
+ A LE R K V +++ L S + DA+ +L + ++ EF+ + LW
Sbjct 29 DGAGLEA-REEKVVYSRSQLSLADSTKALGDAF---------KLFMPRSTEFMSSDAELW 78
Query 81 KEACSLEDERNARILLTRAVECVS 104
CSL+ + + IL ++ V S
Sbjct 79 SFLCSLKHQFSPHILRSKDVYGYS 102
> ath:AT3G13210 crooked neck protein, putative / cell cycle protein,
putative
Length=657
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 34/72 (47%), Gaps = 5/72 (6%)
Query 49 WLDAYAREKDINDRRLVLRKALEFIPNSVRLWKEACSLEDE----RNARILLTRAVECVS 104
+ D + K +N+ R V +A+ +P +LW + +E++ AR +L R + C S
Sbjct 97 YADFEMKNKSVNEARNVWDRAVSLLPRVDQLWYKFIHMEEKLGNIAGARQILERWIHC-S 155
Query 105 QSHEMWLALARL 116
+ WL +
Sbjct 156 PDQQAWLCFIKF 167
> xla:446406 ttc37, MGC83808; tetratricopeptide repeat domain
37; K12600 superkiller protein 3
Length=1564
Score = 27.7 bits (60), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 33/74 (44%), Gaps = 7/74 (9%)
Query 18 VWLEAARLEKPRNAKAVLAKAVSVLPHSVRLWLDAYAREKDINDRRL------VLRKALE 71
+ L A+ LE+P A+A KAV + P + W + +N + V +K LE
Sbjct 46 IGLAASELEQPDQAQAAYRKAVEIEPDQLLAWQGLGNLYEKVNQKDFKEDLPNVYQKLLE 105
Query 72 FIPNSVRL-WKEAC 84
+S + W E C
Sbjct 106 LYRSSDKQKWYEIC 119
> ath:AT4G28200 hypothetical protein; K14557 U3 small nucleolar
RNA-associated protein 6
Length=648
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 1 ARELISLGCQRCPKSEDVWLEAARLE 26
AR L+ G + C SED+W+E R+E
Sbjct 164 ARALMLNGLRVCSNSEDLWVEYLRME 189
Lambda K H
0.320 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2008132680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40