bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2964_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
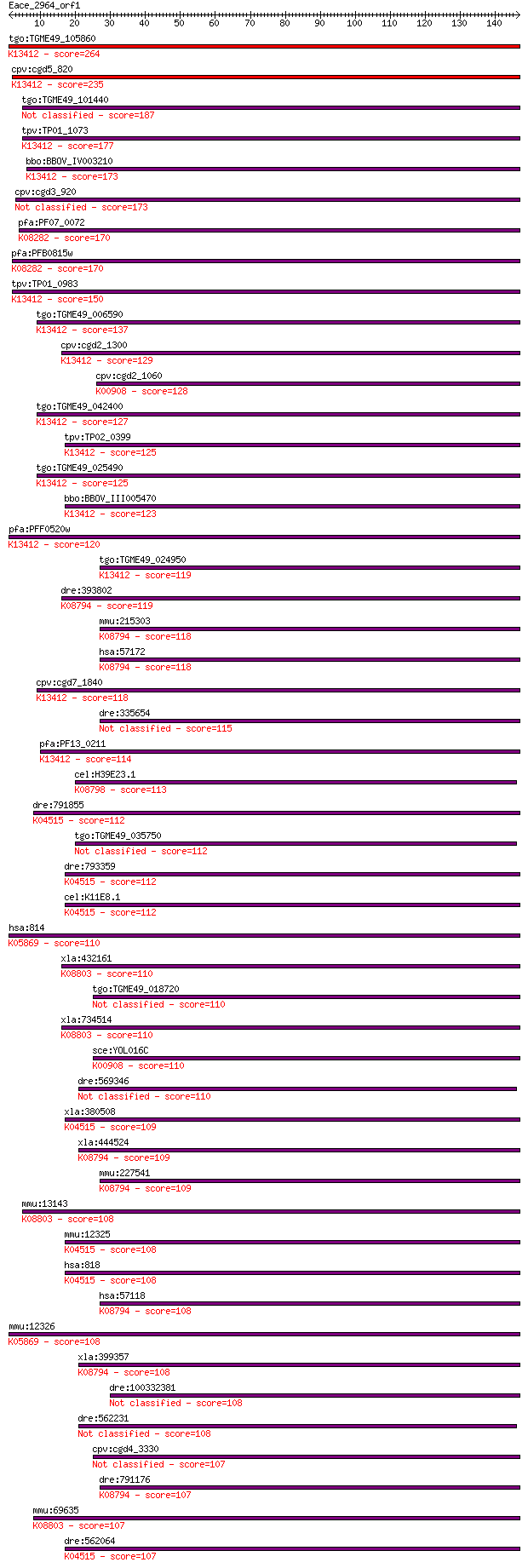
tgo:TGME49_105860 calcium-dependent protein kinase 1, putative... 264 7e-71
cpv:cgd5_820 calcium/calmodulin dependent protein kinase with ... 235 3e-62
tgo:TGME49_101440 calmodulin-domain protein kinase 1, putative... 187 9e-48
tpv:TP01_1073 calmodulin-domain protein kinase; K13412 calcium... 177 1e-44
bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase ... 173 2e-43
cpv:cgd3_920 calmodulin-domain protein kinase 1 173 2e-43
pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:... 170 1e-42
pfa:PFB0815w CDPK1, CPK; Calcium-dependent protein kinase 1 (E... 170 1e-42
tpv:TP01_0983 calcium-dependent protein kinase; K13412 calcium... 150 2e-36
tgo:TGME49_006590 calcium-dependent protein kinase, putative (... 137 1e-32
cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with... 129 3e-30
cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with... 128 6e-30
tgo:TGME49_042400 calcium-dependent protein kinase, putative (... 127 1e-29
tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium... 125 4e-29
tgo:TGME49_025490 calcium-dependent protein kinase, putative (... 125 4e-29
bbo:BBOV_III005470 17.m07489; protein kinase domain containing... 123 2e-28
pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7... 120 1e-27
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 119 2e-27
dre:393802 MGC73155, wu:fk61c03; zgc:73155 (EC:2.7.11.17); K08... 119 3e-27
mmu:215303 Camk1g, CLICK-III, CaMKIgamma, MGC30513; calcium/ca... 118 6e-27
hsa:57172 CAMK1G, CLICKIII, VWS1, dJ272L16.1; calcium/calmodul... 118 6e-27
cpv:cgd7_1840 calcium/calmodulin-dependent protein kinase with... 118 7e-27
dre:335654 camk1g, wu:fk53c02, zgc:73127; calcium/calmodulin-d... 115 6e-26
pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2... 114 1e-25
cel:H39E23.1 par-1; abnormal embryonic PARtitioning of cytopla... 113 2e-25
dre:791855 camk2g1, MGC109908, camk2ga, zgc:109908; calcium/ca... 112 3e-25
tgo:TGME49_035750 calcium/calmodulin-dependent protein kinase ... 112 4e-25
dre:793359 camk2g2; calcium/calmodulin-dependent protein kinas... 112 5e-25
cel:K11E8.1 unc-43; UNCoordinated family member (unc-43); K045... 112 5e-25
hsa:814 CAMK4, CaMK_IV, CaMK-GR, IV, MGC36771, caMK; calcium/c... 110 1e-24
xla:432161 hypothetical protein MGC81183; K08803 death-associa... 110 2e-24
tgo:TGME49_018720 calcium-dependent protein kinase, putative (... 110 2e-24
xla:734514 dapk3, MGC114871; death-associated protein kinase 3... 110 2e-24
sce:YOL016C CMK2; Calmodulin-dependent protein kinase; may pla... 110 2e-24
dre:569346 MAP/microtubule affinity-regulating kinase 3-like 110 2e-24
xla:380508 camk2g, MGC52524, camk2; calcium/calmodulin-depende... 109 3e-24
xla:444524 MGC82022 protein; K08794 calcium/calmodulin-depende... 109 4e-24
mmu:227541 Camk1d, A630059D12Rik, CKLiK, CaMKIdelta, E030025C1... 109 4e-24
mmu:13143 Dapk2; death-associated protein kinase 2 (EC:2.7.11.... 108 5e-24
mmu:12325 Camk2g, 5930429P18Rik, Camkg; calcium/calmodulin-dep... 108 5e-24
hsa:818 CAMK2G, CAMK, CAMK-II, CAMKG, FLJ16043, MGC26678; calc... 108 5e-24
hsa:57118 CAMK1D, CKLiK, CaM-K1, CaMKID; calcium/calmodulin-de... 108 6e-24
mmu:12326 Camk4, A430110E23Rik, AI666733, CaMKIV, CaMKIV/Gr, D... 108 7e-24
xla:399357 camk1d, cam-ki; calcium/calmodulin-dependent protei... 108 7e-24
dre:100332381 calcium/calmodulin-dependent protein kinase IV-like 108 8e-24
dre:562231 fc75f05, wu:fc75f05; si:dkey-175m17.8 108 8e-24
cpv:cgd4_3330 hypothetical protein 107 1e-23
dre:791176 camk1d, zgc:158713; calcium/calmodulin-dependent pr... 107 1e-23
mmu:69635 Dapk1, 2310039H24Rik, 2810425C21Rik, AI642003, D13Uc... 107 1e-23
dre:562064 MGC165373; zgc:165373; K04515 calcium/calmodulin-de... 107 1e-23
> tgo:TGME49_105860 calcium-dependent protein kinase 1, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=537
Score = 264 bits (675), Expect = 7e-71, Method: Compositional matrix adjust.
Identities = 130/147 (88%), Positives = 135/147 (91%), Gaps = 1/147 (0%)
Query 1 TMTMTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSA- 59
+M MTPGMYI QQ+ HLSDRYQRVKKLGSGAYGEVLLCKDK TGAE AIKIIKKSSV+
Sbjct 56 SMPMTPGMYITQQKAHLSDRYQRVKKLGSGAYGEVLLCKDKLTGAERAIKIIKKSSVTTT 115
Query 60 PNSGALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVD 119
NSGALL+EVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVY GGELFDEII RQKFSEVD
Sbjct 116 SNSGALLDEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYRGGELFDEIILRQKFSEVD 175
Query 120 AAVIMKQVLSGVVYLHKHSIVHRDLKP 146
AAVIMKQVLSG YLHKH+IVHRDLKP
Sbjct 176 AAVIMKQVLSGTTYLHKHNIVHRDLKP 202
> cpv:cgd5_820 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=523
Score = 235 bits (600), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 112/145 (77%), Positives = 126/145 (86%), Gaps = 0/145 (0%)
Query 2 MTMTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPN 61
+ TPGM+I ++GHLS+ YQRVKKLGSGAYGEVLLC+DK T E AIKII+K+SVS +
Sbjct 43 LQATPGMFITSKKGHLSEMYQRVKKLGSGAYGEVLLCRDKVTHVERAIKIIRKTSVSTSS 102
Query 62 SGALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAA 121
+ LLEEVAVLK LDHPNIMKLY+FFEDKRNYYLVME Y GGELFDEII R KF+EVDAA
Sbjct 103 NSKLLEEVAVLKLLDHPNIMKLYDFFEDKRNYYLVMECYKGGELFDEIIHRMKFNEVDAA 162
Query 122 VIMKQVLSGVVYLHKHSIVHRDLKP 146
VI+KQVLSGV YLHKH+IVHRDLKP
Sbjct 163 VIIKQVLSGVTYLHKHNIVHRDLKP 187
> tgo:TGME49_101440 calmodulin-domain protein kinase 1, putative
(EC:2.7.11.17)
Length=582
Score = 187 bits (475), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 91/143 (63%), Positives = 111/143 (77%), Gaps = 1/143 (0%)
Query 5 TPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSV-SAPNSG 63
TPGM++ SDRY+ + LG G++GEV+LCKDK TG E A+K+I K V +
Sbjct 35 TPGMFVQHSTAIFSDRYKGQRVLGKGSFGEVILCKDKITGQECAVKVISKRQVKQKTDKE 94
Query 64 ALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVI 123
+LL EV +LKQLDHPNIMKLYEFFEDK +YLV EVY GGELFDEIISR++FSEVDAA I
Sbjct 95 SLLREVQLLKQLDHPNIMKLYEFFEDKGYFYLVGEVYTGGELFDEIISRKRFSEVDAARI 154
Query 124 MKQVLSGVVYLHKHSIVHRDLKP 146
++QVLSG+ Y+HK+ IVHRDLKP
Sbjct 155 IRQVLSGITYMHKNKIVHRDLKP 177
> tpv:TP01_1073 calmodulin-domain protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=504
Score = 177 bits (448), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 83/143 (58%), Positives = 109/143 (76%), Gaps = 1/143 (0%)
Query 5 TPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAP-NSG 63
TPGM++ + + + Y+ VK LG G++GEVLLC + TG AIK+I KSS+ +
Sbjct 34 TPGMFVRRSKASFKENYKAVKLLGKGSFGEVLLCIHRVTGIRYAIKVICKSSMKKKGDHD 93
Query 64 ALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVI 123
ALL EV VLK LDHPNIMK++EFFED++ YY V E+Y GGELFDEI+SR++ +E DAA I
Sbjct 94 ALLREVDVLKSLDHPNIMKIFEFFEDEKYYYFVTELYTGGELFDEIVSRKRLNEYDAARI 153
Query 124 MKQVLSGVVYLHKHSIVHRDLKP 146
+KQ+LSG+ Y+HKH+IVHRDLKP
Sbjct 154 IKQILSGITYMHKHNIVHRDLKP 176
> bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase
4 (EC:2.7.11.1); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=517
Score = 173 bits (438), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 83/142 (58%), Positives = 107/142 (75%), Gaps = 1/142 (0%)
Query 6 PGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAP-NSGA 64
PGM++ G + Y+ VK LG G++GEVLLC ++ TG + A+K+I KSSV + +
Sbjct 49 PGMFVTCATGTFRENYKAVKLLGKGSFGEVLLCLNRATGQQYAVKVIIKSSVKRKGDHES 108
Query 65 LLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIM 124
LL EV VLK LDHPNIMK++EFFED++ YY V E+Y GGELFDEI+SR+ FSE DAA I+
Sbjct 109 LLREVNVLKALDHPNIMKIFEFFEDEKYYYFVTELYSGGELFDEIVSRKCFSEHDAAKII 168
Query 125 KQVLSGVVYLHKHSIVHRDLKP 146
KQVLSG+ Y+H +IVHRDLKP
Sbjct 169 KQVLSGINYMHNRNIVHRDLKP 190
> cpv:cgd3_920 calmodulin-domain protein kinase 1
Length=538
Score = 173 bits (438), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 79/144 (54%), Positives = 107/144 (74%), Gaps = 0/144 (0%)
Query 3 TMTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNS 62
++T GM++ G ++RY V LG G++GEVL CKD+ T E A+K+I K+S ++
Sbjct 58 SVTTGMFVQSGSGTFAERYNIVCMLGKGSFGEVLKCKDRITQQEYAVKVINKASAKNKDT 117
Query 63 GALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAV 122
+L EV +LK+LDHPNIMKL+E ED ++Y+V E+Y GGELFDEII R++FSE DAA
Sbjct 118 STILREVELLKKLDHPNIMKLFEILEDSSSFYIVGELYTGGELFDEIIKRKRFSEHDAAR 177
Query 123 IMKQVLSGVVYLHKHSIVHRDLKP 146
I+KQV SG+ Y+HKH+IVHRDLKP
Sbjct 178 IIKQVFSGITYMHKHNIVHRDLKP 201
> pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:2.7.1.11);
K08282 non-specific serine/threonine protein kinase
[EC:2.7.11.1]
Length=528
Score = 170 bits (431), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 82/144 (56%), Positives = 108/144 (75%), Gaps = 1/144 (0%)
Query 4 MTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVS-APNS 62
+ PGM+I +++Y+ +K LG G++GEV+L +DK TG E AIK+I K V +
Sbjct 53 LRPGMFIQNSNVVFNEQYKGIKILGKGSFGEVILSRDKHTGHEYAIKVISKKHVKRKTDK 112
Query 63 GALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAV 122
+LL EV +LK LDH NIMKLYEFFED YYLV +VY GGELFDEIISR++F E+DAA
Sbjct 113 ESLLREVELLKMLDHINIMKLYEFFEDNNYYYLVSDVYTGGELFDEIISRKRFYEIDAAR 172
Query 123 IMKQVLSGVVYLHKHSIVHRDLKP 146
I+KQ+LSG+ Y+HK+++VHRDLKP
Sbjct 173 IIKQILSGITYMHKNNVVHRDLKP 196
> pfa:PFB0815w CDPK1, CPK; Calcium-dependent protein kinase 1
(EC:2.7.11.1); K08282 non-specific serine/threonine protein
kinase [EC:2.7.11.1]
Length=524
Score = 170 bits (430), Expect = 1e-42, Method: Composition-based stats.
Identities = 82/158 (51%), Positives = 110/158 (69%), Gaps = 13/158 (8%)
Query 2 MTMTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPN 61
+ + PGMY+ ++ G + + Y +V+KLGSGAYGEVLLC++K E AIK+IKKS
Sbjct 37 LAINPGMYVRKKEGKIGESYFKVRKLGSGAYGEVLLCREKHGHGEKAIKVIKKSQFDKMK 96
Query 62 SG-------------ALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDE 108
+ E+++LK LDHPNI+KL++ FEDK+ +YLV E Y GGELF++
Sbjct 97 YSITNKIECDDKIHEEIYNEISLLKSLDHPNIIKLFDVFEDKKYFYLVTEFYEGGELFEQ 156
Query 109 IISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
II+R KF E DAA IMKQ+LSG+ YLHKH+IVHRD+KP
Sbjct 157 IINRHKFDECDAANIMKQILSGICYLHKHNIVHRDIKP 194
> tpv:TP01_0983 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=509
Score = 150 bits (378), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 75/146 (51%), Positives = 99/146 (67%), Gaps = 1/146 (0%)
Query 2 MTMTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKK-SSVSAP 60
+ + P MYI + + Y +KLG+GAYG V LC + E A+K+IKK S S
Sbjct 35 IKLKPSMYIGNIKDEFNSLYTVCRKLGTGAYGTVFLCTRINSTKEYAVKVIKKLRSSSID 94
Query 61 NSGALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDA 120
+S L EV LK+LDHPNI +LY+ +ED YY+VME GGELFDEII RQ+ +E ++
Sbjct 95 SSQGRLREVKYLKELDHPNIARLYDVYEDSVAYYIVMEPCYGGELFDEIIKRQRITEHES 154
Query 121 AVIMKQVLSGVVYLHKHSIVHRDLKP 146
A ++KQ+LSGV YLHK++IVHRDLKP
Sbjct 155 ACLIKQILSGVCYLHKNNIVHRDLKP 180
> tgo:TGME49_006590 calcium-dependent protein kinase, putative
(EC:2.7.11.17 3.4.22.53); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=761
Score = 137 bits (344), Expect = 1e-32, Method: Composition-based stats.
Identities = 70/139 (50%), Positives = 93/139 (66%), Gaps = 3/139 (2%)
Query 9 YIVQQRGHLSDRYQ-RVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLE 67
+I++ G L+D Y+ LG G YG V + K TG A+K I KS V N +
Sbjct 212 FILENTGALTDFYEIDTATLGQGTYGSVSKARKKDTGQMRAVKTISKSQVK--NLERFRQ 269
Query 68 EVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQV 127
E+A++K+LDHPN++KL+E FED RN YLVME+ GGELFD IIS + +E AAV+MKQ+
Sbjct 270 EIAIMKELDHPNVIKLFETFEDHRNIYLVMELCTGGELFDRIISEGRLTEKQAAVLMKQM 329
Query 128 LSGVVYLHKHSIVHRDLKP 146
S V YLH ++I+HRDLKP
Sbjct 330 FSAVHYLHSNNIMHRDLKP 348
> cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=677
Score = 129 bits (325), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 66/132 (50%), Positives = 88/132 (66%), Gaps = 3/132 (2%)
Query 16 HLSDRYQ-RVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQ 74
+++D Y+ + LG G+YG V+ DKQ+GA+ A+KII K + N L E+ ++K+
Sbjct 179 NINDFYELNLGNLGRGSYGSVVKAIDKQSGAQRAVKIILKPKLE--NINRLKREILIMKR 236
Query 75 LDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYL 134
LDHPNI+KL+E FED Y VME+ GGELFD II R FSE AAVIM+QV S + Y
Sbjct 237 LDHPNIIKLFEVFEDTNYLYFVMEICTGGELFDRIIKRGHFSERYAAVIMRQVFSAIAYC 296
Query 135 HKHSIVHRDLKP 146
H + +HRDLKP
Sbjct 297 HSNEFMHRDLKP 308
> cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with
a kinas domain and 4 calmodulin-like EF hands ; K00908 Ca2+/calmodulin-dependent
protein kinase [EC:2.7.11.17]
Length=718
Score = 128 bits (322), Expect = 6e-30, Method: Composition-based stats.
Identities = 64/121 (52%), Positives = 83/121 (68%), Gaps = 2/121 (1%)
Query 26 KLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYE 85
++G G YG V ++ TG AIK I + V A ++ ++E+ +LK LDHPNI+KLYE
Sbjct 206 RIGKGTYGSVKSGTNRLTGCIRAIKTIPLTRVEALDN--FMKEINILKNLDHPNIVKLYE 263
Query 86 FFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLK 145
++DK N YLVME+ GGELFD IIS+ F E+ AA +MKQVLS + Y H H IVHRDLK
Sbjct 264 TYQDKENIYLVMELCSGGELFDRIISQGSFDEIYAANLMKQVLSTICYCHDHGIVHRDLK 323
Query 146 P 146
P
Sbjct 324 P 324
> tgo:TGME49_042400 calcium-dependent protein kinase, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=604
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 66/139 (47%), Positives = 87/139 (62%), Gaps = 3/139 (2%)
Query 9 YIVQQRGHLSDRYQ-RVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLE 67
+I+ G L+D Y KKLG G YG V +K T A+K I K+ V N +
Sbjct 116 FILSYSGPLTDYYDVETKKLGQGTYGSVCRAVNKATKNVRAVKTIPKAKVK--NIKRFKQ 173
Query 68 EVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQV 127
E+A++K LDHPNI+KLYE FED +N YLV+E+ GGELFD II FSE+ A +M+Q
Sbjct 174 EIAIMKCLDHPNIIKLYETFEDHKNIYLVLELCKGGELFDRIIEEGYFSEMYAGTLMRQA 233
Query 128 LSGVVYLHKHSIVHRDLKP 146
+ + Y+H+H I HRDLKP
Sbjct 234 FAALYYIHQHGIAHRDLKP 252
> tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=844
Score = 125 bits (314), Expect = 4e-29, Method: Composition-based stats.
Identities = 65/131 (49%), Positives = 89/131 (67%), Gaps = 3/131 (2%)
Query 17 LSDRYQ-RVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQL 75
+ D Y+ + +KLG+G+YG VL K++GA AIKII KS + N+ + E+ ++K L
Sbjct 395 IHDVYEIKSEKLGNGSYGNVLKGVHKESGAVRAIKIILKSKIE--NAMRMKREIQIMKTL 452
Query 76 DHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLH 135
DHPNI+KL+E +ED YLVME+ GGELFD I+S FSE AA IM+QV S + Y H
Sbjct 453 DHPNIIKLFEVYEDADCLYLVMEMCVGGELFDRIVSTNGFSEAYAASIMRQVFSAIAYCH 512
Query 136 KHSIVHRDLKP 146
+++HRDLKP
Sbjct 513 NRNVLHRDLKP 523
> tgo:TGME49_025490 calcium-dependent protein kinase, putative
(EC:1.6.3.1 2.7.11.17 3.2.1.60); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=711
Score = 125 bits (314), Expect = 4e-29, Method: Composition-based stats.
Identities = 61/138 (44%), Positives = 86/138 (62%), Gaps = 2/138 (1%)
Query 9 YIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEE 68
+I+ G +++ Y K +G G +GEV L D TGA A K I K V ++ +E
Sbjct 228 FILANTGPITNYYTVSKTIGRGTWGEVKLVIDNGTGARRAAKKIPKCYVE--DADRFRQE 285
Query 69 VAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVL 128
+ ++K LDHPNI++LYE FED ++YLVME GGELFD ++ + F+E A IM+Q+L
Sbjct 286 IEIMKSLDHPNIVRLYETFEDMTDFYLVMEYCTGGELFDRLVHQGVFTEALACRIMRQIL 345
Query 129 SGVVYLHKHSIVHRDLKP 146
+ V Y H H + HRDLKP
Sbjct 346 AAVAYCHAHRVAHRDLKP 363
> bbo:BBOV_III005470 17.m07489; protein kinase domain containing
protein; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=755
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 60/131 (45%), Positives = 88/131 (67%), Gaps = 3/131 (2%)
Query 17 LSDRYQ-RVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQL 75
++D Y +LG G+YG+VL K+TG A+K+I+K+++ N+ + E+ ++K L
Sbjct 295 ITDVYDLHTNRLGKGSYGQVLKACHKETGEVKAVKVIRKAAIE--NAMRMKREITIMKNL 352
Query 76 DHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLH 135
DHPNI+KL E +ED+ YLVME+ GGELF+EI+ R FSE AA +M+Q+ S + Y H
Sbjct 353 DHPNIVKLLEIYEDEECLYLVMEMCSGGELFEEIVRRGCFSEQYAATMMRQLFSAIAYCH 412
Query 136 KHSIVHRDLKP 146
I+HRDLKP
Sbjct 413 GKGILHRDLKP 423
> pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7.11.1);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=509
Score = 120 bits (302), Expect = 1e-27, Method: Composition-based stats.
Identities = 67/146 (45%), Positives = 87/146 (59%), Gaps = 2/146 (1%)
Query 1 TMTMTPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAP 60
T T+ I+ G L D+Y +KLG G YG V DK T AIK KK +
Sbjct 52 TTTLERKNLILCHSGKLEDKYIIDEKLGQGTYGCVYKGIDKVTNQLYAIKEEKKDRLK-- 109
Query 61 NSGALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDA 120
N +E+ ++K+LDHPNI+KLYE +E+ YL+ME+ G ELFD II F+E +A
Sbjct 110 NINRFFQEIEIMKKLDHPNIVKLYETYENDNYIYLIMELCSGRELFDSIIENGSFTEKNA 169
Query 121 AVIMKQVLSGVVYLHKHSIVHRDLKP 146
A IMKQ+ S + YLH +IVHRDLKP
Sbjct 170 ATIMKQIFSAIFYLHSLNIVHRDLKP 195
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 79/120 (65%), Gaps = 2/120 (1%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LG G+YG V+ KD +TG AIKI+ K + N L E+ ++K+LDHPNI+KL E
Sbjct 219 LGKGSYGSVVKAKDLKTGIMRAIKIVYKPRID--NVTRLKREILIMKRLDHPNIIKLLEV 276
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+ED +N YLVME+ GGELF+ II FSE AA +MKQV S Y H +++HRDLKP
Sbjct 277 YEDMKNLYLVMELCTGGELFERIIKSGHFSERYAASLMKQVFSAASYCHSQNVIHRDLKP 336
> dre:393802 MGC73155, wu:fk61c03; zgc:73155 (EC:2.7.11.17); K08794
calcium/calmodulin-dependent protein kinase I [EC:2.7.11.17]
Length=433
Score = 119 bits (298), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 58/131 (44%), Positives = 90/131 (68%), Gaps = 3/131 (2%)
Query 16 HLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQL 75
++ D + + LGSGA+ EV + K+++TG A+K +KK + N L E+AVL+++
Sbjct 16 NIQDVFDFMDVLGSGAFSEVFMVKERKTGKLFAMKCVKKKNKRDIN---LENEIAVLRKI 72
Query 76 DHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLH 135
H N++ L +F+E + +YYLVM++ GGELFD I+ R +SE+DA+ +++QVL V YLH
Sbjct 73 KHENVVCLEDFYESRTHYYLVMQLVSGGELFDRILDRGMYSEMDASSVIRQVLEAVSYLH 132
Query 136 KHSIVHRDLKP 146
+ IVHRDLKP
Sbjct 133 NNGIVHRDLKP 143
> mmu:215303 Camk1g, CLICK-III, CaMKIgamma, MGC30513; calcium/calmodulin-dependent
protein kinase I gamma (EC:2.7.11.17);
K08794 calcium/calmodulin-dependent protein kinase I [EC:2.7.11.17]
Length=477
Score = 118 bits (296), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 84/120 (70%), Gaps = 2/120 (1%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LGSGA+ EV L K + TG A+K IKKS A +L E+AVLK++ H NI+ L +
Sbjct 29 LGSGAFSEVFLVKQRVTGKLFALKCIKKSP--AFRDSSLENEIAVLKRIKHENIVTLEDI 86
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+E +YYLVM++ GGELFD I+ R ++E DA+++++QVLS V YLH++ IVHRDLKP
Sbjct 87 YESTTHYYLVMQLVSGGELFDRILERGVYTEKDASLVIQQVLSAVKYLHENGIVHRDLKP 146
> hsa:57172 CAMK1G, CLICKIII, VWS1, dJ272L16.1; calcium/calmodulin-dependent
protein kinase IG (EC:2.7.11.17); K08794 calcium/calmodulin-dependent
protein kinase I [EC:2.7.11.17]
Length=476
Score = 118 bits (296), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 84/120 (70%), Gaps = 2/120 (1%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LGSGA+ EV L K + TG A+K IKKS A +L E+AVLK++ H NI+ L +
Sbjct 29 LGSGAFSEVFLVKQRLTGKLFALKCIKKSP--AFRDSSLENEIAVLKKIKHENIVTLEDI 86
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+E +YYLVM++ GGELFD I+ R ++E DA+++++QVLS V YLH++ IVHRDLKP
Sbjct 87 YESTTHYYLVMQLVSGGELFDRILERGVYTEKDASLVIQQVLSAVKYLHENGIVHRDLKP 146
> cpv:cgd7_1840 calcium/calmodulin-dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=676
Score = 118 bits (295), Expect = 7e-27, Method: Composition-based stats.
Identities = 60/138 (43%), Positives = 85/138 (61%), Gaps = 2/138 (1%)
Query 9 YIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEE 68
+I+ +G ++ Y +G G++GEV + K T A K I K V + +E
Sbjct 194 FILSTKGDINQYYTLENTIGRGSWGEVKIAVQKGTRIRRAAKKIPKYFVE--DVDRFKQE 251
Query 69 VAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVL 128
+ ++K LDHPNI++LYE FED + YLVME+ GGELF+ ++ ++ F E DAA IMK VL
Sbjct 252 IEIMKSLDHPNIIRLYETFEDNTDIYLVMELCTGGELFERVVHKRVFRESDAARIMKDVL 311
Query 129 SGVVYLHKHSIVHRDLKP 146
S V Y HK ++ HRDLKP
Sbjct 312 SAVAYCHKLNVAHRDLKP 329
> dre:335654 camk1g, wu:fk53c02, zgc:73127; calcium/calmodulin-dependent
protein kinase IG (EC:2.7.11.17)
Length=426
Score = 115 bits (287), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 56/120 (46%), Positives = 84/120 (70%), Gaps = 3/120 (2%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LGSG++ EV L +++++G A+K +KK + N L E+ VLK++ H N++ L +F
Sbjct 27 LGSGSFSEVYLVRERKSGNFYALKCVKKKQLHHSN---LENEIQVLKRIKHSNVVGLEDF 83
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+E + +YYLVME+ GGELFD I+ R ++E DA+ ++ QVL V YLH++SIVHRDLKP
Sbjct 84 YESRTHYYLVMELVSGGELFDRILDRGVYTEKDASRVINQVLEAVSYLHQNSIVHRDLKP 143
> pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=568
Score = 114 bits (285), Expect = 1e-25, Method: Composition-based stats.
Identities = 66/138 (47%), Positives = 90/138 (65%), Gaps = 3/138 (2%)
Query 10 IVQQRGHLSDRYQRVK-KLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEE 68
I+ LSD Y+ + KLG G+YG V+ K+TG + AIKII+K + N L E
Sbjct 113 ILMNNDELSDVYEIDRYKLGKGSYGNVVKAVSKRTGQQRAIKIIEKKKIH--NIERLKRE 170
Query 69 VAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVL 128
+ ++KQ+DHPNI+KLYE +ED YLV+E+ GGELFD+I+ FSE +A IMKQ+
Sbjct 171 ILIMKQMDHPNIIKLYEVYEDNEKLYLVLELCDGGELFDKIVKYGSFSEYEAYKIMKQIF 230
Query 129 SGVVYLHKHSIVHRDLKP 146
S + Y H +I+HRDLKP
Sbjct 231 SALYYCHSKNIMHRDLKP 248
> cel:H39E23.1 par-1; abnormal embryonic PARtitioning of cytoplasm
family member (par-1); K08798 MAP/microtubule affinity-regulating
kinase [EC:2.7.11.1]
Length=1192
Score = 113 bits (283), Expect = 2e-25, Method: Composition-based stats.
Identities = 54/126 (42%), Positives = 83/126 (65%), Gaps = 0/126 (0%)
Query 20 RYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPN 79
+Y+ +K +G G + +V L K TG E AIKII K++++ + L EV ++KQLDHPN
Sbjct 169 KYKLLKTIGKGNFAKVKLAKHVITGHEVAIKIIDKTALNPSSLQKLFREVKIMKQLDHPN 228
Query 80 IMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSI 139
I+KLY+ E ++ YLV+E GGE+FD +++ + E +A +Q++S V YLH +I
Sbjct 229 IVKLYQVMETEQTLYLVLEYASGGEVFDYLVAHGRMKEKEARAKFRQIVSAVQYLHSKNI 288
Query 140 VHRDLK 145
+HRDLK
Sbjct 289 IHRDLK 294
> dre:791855 camk2g1, MGC109908, camk2ga, zgc:109908; calcium/calmodulin-dependent
protein kinase (CaM kinase) II gamma 1;
K04515 calcium/calmodulin-dependent protein kinase (CaM kinase)
II [EC:2.7.11.17]
Length=560
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 55/139 (39%), Positives = 85/139 (61%), Gaps = 0/139 (0%)
Query 8 MYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLE 67
M + +D YQ ++LG GA+ V C K TG E A KII +SA + L
Sbjct 1 MATIVTSTRFTDEYQLYEELGKGAFSVVRRCVKKSTGQEYAAKIINTKKLSARDHQKLER 60
Query 68 EVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQV 127
E + + L HPNI++L++ ++ +YLV ++ GGELF++I++R+ +SE DA+ + Q+
Sbjct 61 EARICRLLKHPNIVRLHDSISEEGFHYLVFDLVTGGELFEDIVAREYYSEADASHCISQI 120
Query 128 LSGVVYLHKHSIVHRDLKP 146
L V ++H+H IVHRDLKP
Sbjct 121 LESVNHIHQHDIVHRDLKP 139
> tgo:TGME49_035750 calcium/calmodulin-dependent protein kinase
type I, putative (EC:2.7.11.17)
Length=376
Score = 112 bits (280), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 57/135 (42%), Positives = 87/135 (64%), Gaps = 9/135 (6%)
Query 20 RYQRVKK-LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVS----AP----NSGALLEEVA 70
R+ R+ + LGSG++G+V C ++TG A+KII++ + AP N + EV
Sbjct 25 RHYRIGRVLGSGSFGQVRECTRRETGEVFAVKIIERKMTAKEQMAPGRPSNEAMIKSEVN 84
Query 71 VLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSG 130
+LK LDHPN++K +FFED+ +Y V+E+ GGELF EI+ R+ +E DA+ KQ++
Sbjct 85 ILKDLDHPNVVKYVDFFEDRHFFYAVLELCDGGELFHEIVKRRHVTEQDASNFCKQMVGA 144
Query 131 VVYLHKHSIVHRDLK 145
+ YLH+ IVHRD+K
Sbjct 145 LAYLHERGIVHRDVK 159
> dre:793359 camk2g2; calcium/calmodulin-dependent protein kinase
(CaM kinase) II gamma 2; K04515 calcium/calmodulin-dependent
protein kinase (CaM kinase) II [EC:2.7.11.17]
Length=557
Score = 112 bits (280), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/130 (41%), Positives = 83/130 (63%), Gaps = 0/130 (0%)
Query 17 LSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLD 76
+D YQ ++LG GA+ V C K TG E A KII +SA + L E + + L
Sbjct 10 FTDEYQLYEELGKGAFSVVRRCVKKSTGQEYAAKIINTKKLSARDHQKLEREARICRLLK 69
Query 77 HPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHK 136
HPNI++L++ ++ +YLV ++ GGELF++I++R+ +SE DA+ + Q+L V ++H+
Sbjct 70 HPNIVRLHDSIAEEGFHYLVFDLVTGGELFEDIVAREYYSESDASHCINQILESVSHIHQ 129
Query 137 HSIVHRDLKP 146
H IVHRDLKP
Sbjct 130 HDIVHRDLKP 139
> cel:K11E8.1 unc-43; UNCoordinated family member (unc-43); K04515
calcium/calmodulin-dependent protein kinase (CaM kinase)
II [EC:2.7.11.17]
Length=518
Score = 112 bits (279), Expect = 5e-25, Method: Composition-based stats.
Identities = 53/130 (40%), Positives = 83/130 (63%), Gaps = 0/130 (0%)
Query 17 LSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLD 76
SD Y ++LG GA+ V C K TG E A KII +SA + L E + ++L
Sbjct 8 FSDNYDVKEELGKGAFSVVRRCVHKTTGLEFAAKIINTKKLSARDFQKLEREARICRKLQ 67
Query 77 HPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHK 136
HPNI++L++ +++ +YLV ++ GGELF++I++R+ +SE DA+ ++Q+L + Y H
Sbjct 68 HPNIVRLHDSIQEESFHYLVFDLVTGGELFEDIVAREFYSEADASHCIQQILESIAYCHS 127
Query 137 HSIVHRDLKP 146
+ IVHRDLKP
Sbjct 128 NGIVHRDLKP 137
> hsa:814 CAMK4, CaMK_IV, CaMK-GR, IV, MGC36771, caMK; calcium/calmodulin-dependent
protein kinase IV (EC:2.7.11.17); K05869
calcium/calmodulin-dependent protein kinase IV [EC:2.7.11.17]
Length=473
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 63/147 (42%), Positives = 87/147 (59%), Gaps = 5/147 (3%)
Query 1 TMTMTPGMYIV-QQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSA 59
T ++ P +I R LSD ++ +LG GA V CK K T A+K++KK+
Sbjct 25 TASLVPDYWIDGSNRDALSDFFEVESELGRGATSIVYRCKQKGTQKPYALKVLKKTV--- 81
Query 60 PNSGALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVD 119
+ + E+ VL +L HPNI+KL E FE LV+E+ GGELFD I+ + +SE D
Sbjct 82 -DKKIVRTEIGVLLRLSHPNIIKLKEIFETPTEISLVLELVTGGELFDRIVEKGYYSERD 140
Query 120 AAVIMKQVLSGVVYLHKHSIVHRDLKP 146
AA +KQ+L V YLH++ IVHRDLKP
Sbjct 141 AADAVKQILEAVAYLHENGIVHRDLKP 167
> xla:432161 hypothetical protein MGC81183; K08803 death-associated
protein kinase [EC:2.7.11.1]
Length=452
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/135 (40%), Positives = 86/135 (63%), Gaps = 4/135 (2%)
Query 16 HLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEE----VAV 71
++ D Y+ ++LGSG + V C+++ TG E A K IKK +S+ G EE V +
Sbjct 8 NVDDFYEMGEELGSGQFAIVRKCRERSTGVEYAAKFIKKRRLSSSRRGVSREEIEREVDI 67
Query 72 LKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGV 131
L+++ HPNI+ L++ FE++ + L++E+ GGELFD + ++ SE +A + +KQ+L GV
Sbjct 68 LREIQHPNIITLHDVFENRTDVVLILELVSGGELFDFLAQKESLSEEEATMFLKQILDGV 127
Query 132 VYLHKHSIVHRDLKP 146
YLH +I H DLKP
Sbjct 128 HYLHHKNIAHFDLKP 142
> tgo:TGME49_018720 calcium-dependent protein kinase, putative
(EC:2.7.11.17)
Length=557
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/126 (45%), Positives = 79/126 (62%), Gaps = 5/126 (3%)
Query 25 KKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLY 84
+KLGSGA+G+V L +++ +G E IK I K P + E+ VLK LDHPNI+K++
Sbjct 121 RKLGSGAFGDVHLVEERSSGLERVIKTINKDRSQVPME-QIEAEIEVLKSLDHPNIIKIF 179
Query 85 EFFEDKRNYYLVMEVYGGGELFDEIISRQ----KFSEVDAAVIMKQVLSGVVYLHKHSIV 140
E FED N Y+VME GGEL + I+S Q SE A +MKQ+++ + Y H +V
Sbjct 180 EVFEDYHNMYIVMETCEGGELLERIVSAQARGKALSEGYVAELMKQMMNALAYFHSQHVV 239
Query 141 HRDLKP 146
H+DLKP
Sbjct 240 HKDLKP 245
> xla:734514 dapk3, MGC114871; death-associated protein kinase
3 (EC:2.7.11.1); K08803 death-associated protein kinase [EC:2.7.11.1]
Length=452
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/135 (40%), Positives = 86/135 (63%), Gaps = 4/135 (2%)
Query 16 HLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEE----VAV 71
++ D Y+ ++LGSG + V C+++ TG E A K IKK +S+ G EE V +
Sbjct 8 NVDDFYEMGEELGSGQFAIVRKCRERSTGVEYAAKFIKKRRLSSSRRGVSREEIEREVDI 67
Query 72 LKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGV 131
L+++ HPNI+ L++ FE++ + L++E+ GGELFD + ++ SE +A + +KQ+L GV
Sbjct 68 LREIQHPNIITLHDVFENRTDVVLILELVSGGELFDFLAQKESLSEEEATMFLKQILDGV 127
Query 132 VYLHKHSIVHRDLKP 146
YLH +I H DLKP
Sbjct 128 HYLHHKNIAHFDLKP 142
> sce:YOL016C CMK2; Calmodulin-dependent protein kinase; may play
a role in stress response, many CA++/calmodulan dependent
phosphorylation substrates demonstrated in vitro, amino acid
sequence similar to Cmk1p and mammalian Cam Kinase II (EC:2.7.11.17);
K00908 Ca2+/calmodulin-dependent protein kinase
[EC:2.7.11.17]
Length=447
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/124 (43%), Positives = 82/124 (66%), Gaps = 2/124 (1%)
Query 25 KKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSG--ALLEEVAVLKQLDHPNIMK 82
+ LG+G++G V + T + AIKI+ K ++ N L EE+++L++L HPNI+
Sbjct 51 RTLGAGSFGVVRQARKLSTNEDVAIKILLKKALQGNNVQLQMLYEELSILQKLSHPNIVS 110
Query 83 LYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHR 142
++FE K +Y+V ++ GGELFD I+SR KF+EVDA I+ Q+L V Y+H ++VHR
Sbjct 111 FKDWFESKDKFYIVTQLATGGELFDRILSRGKFTEVDAVEIIVQILGAVEYMHSKNVVHR 170
Query 143 DLKP 146
DLKP
Sbjct 171 DLKP 174
> dre:569346 MAP/microtubule affinity-regulating kinase 3-like
Length=743
Score = 110 bits (274), Expect = 2e-24, Method: Composition-based stats.
Identities = 52/125 (41%), Positives = 80/125 (64%), Gaps = 0/125 (0%)
Query 21 YQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNI 80
Y+ +K +G G + +V L + TG E AIKII K+ ++ + L EV ++K L HPNI
Sbjct 58 YRLLKTIGKGNFAKVKLARHILTGKEVAIKIIDKTQLNPTSLQKLFREVRIMKTLHHPNI 117
Query 81 MKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIV 140
++L+E E ++ YLVME GGE+FD ++S + E++A +Q++S V Y H+ +IV
Sbjct 118 VQLFEVIETEKTLYLVMEYASGGEVFDYLVSHGRMKEIEARAKFRQIVSAVHYCHQKNIV 177
Query 141 HRDLK 145
HRDLK
Sbjct 178 HRDLK 182
> xla:380508 camk2g, MGC52524, camk2; calcium/calmodulin-dependent
protein kinase (CaM kinase) II gamma (EC:2.7.11.17); K04515
calcium/calmodulin-dependent protein kinase (CaM kinase)
II [EC:2.7.11.17]
Length=564
Score = 109 bits (272), Expect = 3e-24, Method: Composition-based stats.
Identities = 51/130 (39%), Positives = 82/130 (63%), Gaps = 0/130 (0%)
Query 17 LSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLD 76
+D YQ ++LG GA+ V C K + E A KII +SA + L E + + L
Sbjct 10 FTDEYQLYEELGKGAFSVVRRCVKKSSSQEYAAKIINTKKLSARDHQKLEREARICRLLK 69
Query 77 HPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHK 136
HPNI++L++ ++ +YL+ ++ GGELF++I++R+ +SE DA+ + Q+L V ++H+
Sbjct 70 HPNIVRLHDSISEEGFHYLLFDLVTGGELFEDIVAREYYSEADASHCIHQILESVNHIHQ 129
Query 137 HSIVHRDLKP 146
H IVHRDLKP
Sbjct 130 HDIVHRDLKP 139
> xla:444524 MGC82022 protein; K08794 calcium/calmodulin-dependent
protein kinase I [EC:2.7.11.17]
Length=395
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 54/126 (42%), Positives = 86/126 (68%), Gaps = 1/126 (0%)
Query 21 YQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNI 80
+Q + LG+GA+ EV+L ++K+TG A+K I K ++ S ++ E+AVL+++ H NI
Sbjct 24 FQFKEVLGTGAFSEVVLAEEKETGKLFAVKCIPKKALKGKES-SIENEIAVLRKIKHENI 82
Query 81 MKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIV 140
+ L + +E + YLVM++ GGELFD I+ + ++E DA+ +++QVL V YLH+ IV
Sbjct 83 VALEDIYESPSHLYLVMQLVSGGELFDRIVEKGFYTEKDASTLIRQVLDAVSYLHRLGIV 142
Query 141 HRDLKP 146
HRDLKP
Sbjct 143 HRDLKP 148
> mmu:227541 Camk1d, A630059D12Rik, CKLiK, CaMKIdelta, E030025C11Rik;
calcium/calmodulin-dependent protein kinase ID (EC:2.7.11.17);
K08794 calcium/calmodulin-dependent protein kinase
I [EC:2.7.11.17]
Length=385
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 53/120 (44%), Positives = 82/120 (68%), Gaps = 1/120 (0%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LG+GA+ EV+L ++K TG A+K I K ++ S ++ E+AVL+++ H NI+ L +
Sbjct 29 LGTGAFSEVVLAEEKATGKLFAVKCIPKKALKGKES-SIENEIAVLRKIKHENIVALEDI 87
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+E + YLVM++ GGELFD I+ + ++E DA+ +++QVL V YLH+ IVHRDLKP
Sbjct 88 YESPNHLYLVMQLVSGGELFDRIVEKGFYTEKDASTLIRQVLDAVYYLHRMGIVHRDLKP 147
> mmu:13143 Dapk2; death-associated protein kinase 2 (EC:2.7.11.1);
K08803 death-associated protein kinase [EC:2.7.11.1]
Length=370
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 58/146 (39%), Positives = 87/146 (59%), Gaps = 5/146 (3%)
Query 5 TPGMYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGA 64
+P M +Q+ + D Y ++LGSG + V C++K TG E A K IKK A G
Sbjct 8 SPNMETFKQQK-VEDFYDIGEELGSGQFAIVKKCREKSTGLEYAAKFIKKRQSRASRRGV 66
Query 65 LLEE----VAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDA 120
EE V++L+Q+ HPNI+ L++ +E++ + L++E+ GGELFD + ++ SE +A
Sbjct 67 CREEIEREVSILRQVLHPNIITLHDVYENRTDVVLILELVSGGELFDFLAQKESLSEEEA 126
Query 121 AVIMKQVLSGVVYLHKHSIVHRDLKP 146
+KQ+L GV YLH I H DLKP
Sbjct 127 TSFIKQILDGVNYLHTKKIAHFDLKP 152
> mmu:12325 Camk2g, 5930429P18Rik, Camkg; calcium/calmodulin-dependent
protein kinase II gamma (EC:2.7.11.17); K04515 calcium/calmodulin-dependent
protein kinase (CaM kinase) II [EC:2.7.11.17]
Length=518
Score = 108 bits (271), Expect = 5e-24, Method: Composition-based stats.
Identities = 52/130 (40%), Positives = 82/130 (63%), Gaps = 0/130 (0%)
Query 17 LSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLD 76
+D YQ ++LG GA+ V C K + E A KII +SA + L E + + L
Sbjct 10 FTDDYQLFEELGKGAFSVVRRCVKKTSTQEYAAKIINTKKLSARDHQKLEREARICRLLK 69
Query 77 HPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHK 136
HPNI++L++ ++ +YLV ++ GGELF++I++R+ +SE DA+ + Q+L V ++H+
Sbjct 70 HPNIVRLHDSISEEGFHYLVFDLVTGGELFEDIVAREYYSEADASHCIHQILESVNHIHQ 129
Query 137 HSIVHRDLKP 146
H IVHRDLKP
Sbjct 130 HDIVHRDLKP 139
> hsa:818 CAMK2G, CAMK, CAMK-II, CAMKG, FLJ16043, MGC26678; calcium/calmodulin-dependent
protein kinase II gamma (EC:2.7.11.17);
K04515 calcium/calmodulin-dependent protein kinase (CaM
kinase) II [EC:2.7.11.17]
Length=539
Score = 108 bits (270), Expect = 5e-24, Method: Composition-based stats.
Identities = 52/130 (40%), Positives = 82/130 (63%), Gaps = 0/130 (0%)
Query 17 LSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLD 76
+D YQ ++LG GA+ V C K + E A KII +SA + L E + + L
Sbjct 10 FTDDYQLFEELGKGAFSVVRRCVKKTSTQEYAAKIINTKKLSARDHQKLEREARICRLLK 69
Query 77 HPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHK 136
HPNI++L++ ++ +YLV ++ GGELF++I++R+ +SE DA+ + Q+L V ++H+
Sbjct 70 HPNIVRLHDSISEEGFHYLVFDLVTGGELFEDIVAREYYSEADASHCIHQILESVNHIHQ 129
Query 137 HSIVHRDLKP 146
H IVHRDLKP
Sbjct 130 HDIVHRDLKP 139
> hsa:57118 CAMK1D, CKLiK, CaM-K1, CaMKID; calcium/calmodulin-dependent
protein kinase ID (EC:2.7.11.17); K08794 calcium/calmodulin-dependent
protein kinase I [EC:2.7.11.17]
Length=357
Score = 108 bits (270), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 53/120 (44%), Positives = 82/120 (68%), Gaps = 1/120 (0%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LG+GA+ EV+L ++K TG A+K I K ++ S ++ E+AVL+++ H NI+ L +
Sbjct 29 LGTGAFSEVVLAEEKATGKLFAVKCIPKKALKGKES-SIENEIAVLRKIKHENIVALEDI 87
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+E + YLVM++ GGELFD I+ + ++E DA+ +++QVL V YLH+ IVHRDLKP
Sbjct 88 YESPNHLYLVMQLVSGGELFDRIVEKGFYTEKDASTLIRQVLDAVYYLHRMGIVHRDLKP 147
> mmu:12326 Camk4, A430110E23Rik, AI666733, CaMKIV, CaMKIV/Gr,
D18Bwg0362e; calcium/calmodulin-dependent protein kinase IV
(EC:2.7.11.17); K05869 calcium/calmodulin-dependent protein
kinase IV [EC:2.7.11.17]
Length=469
Score = 108 bits (269), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 62/147 (42%), Positives = 85/147 (57%), Gaps = 5/147 (3%)
Query 1 TMTMTPGMYIV-QQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSA 59
T + P +I R L D ++ +LG GA V CK K T A+K++KK+
Sbjct 21 TENLVPDYWIDGSNRDPLGDFFEVESELGRGATSIVYRCKQKGTQKPYALKVLKKTV--- 77
Query 60 PNSGALLEEVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVD 119
+ + E+ VL +L HPNI+KL E FE LV+E+ GGELFD I+ + +SE D
Sbjct 78 -DKKIVRTEIGVLLRLSHPNIIKLKEIFETPTEISLVLELVTGGELFDRIVEKGYYSERD 136
Query 120 AAVIMKQVLSGVVYLHKHSIVHRDLKP 146
AA +KQ+L V YLH++ IVHRDLKP
Sbjct 137 AADAVKQILEAVAYLHENGIVHRDLKP 163
> xla:399357 camk1d, cam-ki; calcium/calmodulin-dependent protein
kinase ID; K08794 calcium/calmodulin-dependent protein kinase
I [EC:2.7.11.17]
Length=395
Score = 108 bits (269), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 54/126 (42%), Positives = 85/126 (67%), Gaps = 1/126 (0%)
Query 21 YQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNI 80
+Q + LG+GA+ EV+L ++K TG A+K I K ++ S ++ E+AVL+++ H NI
Sbjct 24 FQFKEVLGTGAFSEVVLAEEKATGKLFAVKCIPKKALKGKES-SIENEIAVLRKIKHENI 82
Query 81 MKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIV 140
+ L + +E + YLVM++ GGELFD I+ + ++E DA+ +++QVL V YLH+ IV
Sbjct 83 VALEDIYESPSHLYLVMQLVSGGELFDRIVEKGFYTEKDASTLIRQVLDAVSYLHRLGIV 142
Query 141 HRDLKP 146
HRDLKP
Sbjct 143 HRDLKP 148
> dre:100332381 calcium/calmodulin-dependent protein kinase IV-like
Length=406
Score = 108 bits (269), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 53/117 (45%), Positives = 78/117 (66%), Gaps = 4/117 (3%)
Query 30 GAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEFFED 89
GA VL C++KQT A+K++KK+ + + E+ VL +L HPNI++L E FE
Sbjct 5 GATSVVLRCEEKQTEKPYAVKVLKKTI----DKKIVRTEIGVLLRLSHPNIIRLKEIFET 60
Query 90 KRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+ +L++E+ GGELFD I+ R +SE DAA ++KQ+L V YLH++ +VHRDLKP
Sbjct 61 ETEIFLILELVTGGELFDRIVERGYYSERDAAHVIKQILEAVAYLHENGVVHRDLKP 117
> dre:562231 fc75f05, wu:fc75f05; si:dkey-175m17.8
Length=793
Score = 108 bits (269), Expect = 8e-24, Method: Composition-based stats.
Identities = 52/125 (41%), Positives = 80/125 (64%), Gaps = 0/125 (0%)
Query 21 YQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNI 80
Y+ +K +G G + +V L K TG E A+KII K+ +++ + L EV ++K L+HPNI
Sbjct 44 YRLLKTIGKGNFAKVKLAKHILTGKEVAVKIIDKTQLNSSSLQKLFREVRIMKLLNHPNI 103
Query 81 MKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIV 140
+KL+E E ++ YLVME GGE+FD +++ + E +A +Q++S V Y H+ IV
Sbjct 104 VKLFEVIETEKTLYLVMEYASGGEVFDYLVAHGRMKEKEARAKFRQIVSAVQYCHQKCIV 163
Query 141 HRDLK 145
HRDLK
Sbjct 164 HRDLK 168
> cpv:cgd4_3330 hypothetical protein
Length=622
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 56/126 (44%), Positives = 86/126 (68%), Gaps = 5/126 (3%)
Query 25 KKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLY 84
KK+G G++G+V L +++ +G KII +S +S + + EVAVLK LDHPNI+K++
Sbjct 184 KKIGEGSFGKVFLVRERCSGLTRVCKIIDRS-LSNMSLEQIEAEVAVLKSLDHPNIIKIF 242
Query 85 EFFEDKRNYYLVMEVYGGGELFDEI---ISRQ-KFSEVDAAVIMKQVLSGVVYLHKHSIV 140
E +EDK + Y++ME GGGELF+ I +S+ + SE A IM+Q+++ + Y H +IV
Sbjct 243 EVYEDKEHMYIIMENCGGGELFERIHEAVSKGFRLSERYVAHIMRQIMAALAYFHSRNIV 302
Query 141 HRDLKP 146
H+DLKP
Sbjct 303 HKDLKP 308
> dre:791176 camk1d, zgc:158713; calcium/calmodulin-dependent
protein kinase 1D (EC:2.7.11.17); K08794 calcium/calmodulin-dependent
protein kinase I [EC:2.7.11.17]
Length=392
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 54/120 (45%), Positives = 80/120 (66%), Gaps = 1/120 (0%)
Query 27 LGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLDHPNIMKLYEF 86
LG+GA+ EV+L ++K TG A+K I K ++ SG + E+AVL+++ H NI+ L +
Sbjct 30 LGTGAFSEVVLAQEKATGDMYAVKCIPKKALRGKESG-IENEIAVLRKIKHENIVALEDI 88
Query 87 FEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHKHSIVHRDLKP 146
+E + YL+M++ GGELFD I+ R ++E A+ ++KQVL V YLH IVHRDLKP
Sbjct 89 YESPSHLYLIMQLVSGGELFDRIVERGFYTEQGASALIKQVLDAVNYLHSLGIVHRDLKP 148
> mmu:69635 Dapk1, 2310039H24Rik, 2810425C21Rik, AI642003, D13Ucla1,
DAP-Kinase; death associated protein kinase 1 (EC:2.7.11.1);
K08803 death-associated protein kinase [EC:2.7.11.1]
Length=1430
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 56/143 (39%), Positives = 87/143 (60%), Gaps = 5/143 (3%)
Query 8 MYIVQQRGHLSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLE 67
M + +Q ++ D Y ++LGSG + V C++K TG + A K IKK + G E
Sbjct 1 MTVFRQE-NVDDYYDTGEELGSGQFAVVKKCREKSTGLQYAAKFIKKRRTKSSRRGVSRE 59
Query 68 ----EVAVLKQLDHPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVI 123
EV++LK++ HPN++ L+E +E+K + L++E+ GGELFD + ++ +E +A
Sbjct 60 DIEREVSILKEIRHPNVITLHEVYENKTDVILILELVAGGELFDFLAEKESLTEEEATEF 119
Query 124 MKQVLSGVVYLHKHSIVHRDLKP 146
+KQ+LSGV YLH I H DLKP
Sbjct 120 LKQILSGVYYLHSLQIAHFDLKP 142
> dre:562064 MGC165373; zgc:165373; K04515 calcium/calmodulin-dependent
protein kinase (CaM kinase) II [EC:2.7.11.17]
Length=618
Score = 107 bits (267), Expect = 1e-23, Method: Composition-based stats.
Identities = 50/130 (38%), Positives = 82/130 (63%), Gaps = 0/130 (0%)
Query 17 LSDRYQRVKKLGSGAYGEVLLCKDKQTGAEGAIKIIKKSSVSAPNSGALLEEVAVLKQLD 76
+D YQ ++LG GA+ V C TG E A KII +SA + L E + + L
Sbjct 9 FTDEYQLYEELGKGAFSVVRRCVKLSTGQEYAAKIINTKKLSARDHQKLEREARICRLLK 68
Query 77 HPNIMKLYEFFEDKRNYYLVMEVYGGGELFDEIISRQKFSEVDAAVIMKQVLSGVVYLHK 136
HPNI++L++ ++ +YL+ ++ GGELF++I++R+ +SE DA+ ++Q+L V++ H+
Sbjct 69 HPNIVRLHDSISEEGFHYLLFDLVTGGELFEDIVAREYYSEADASHCIQQILEAVLHCHQ 128
Query 137 HSIVHRDLKP 146
+VHRDLKP
Sbjct 129 MGVVHRDLKP 138
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40