bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
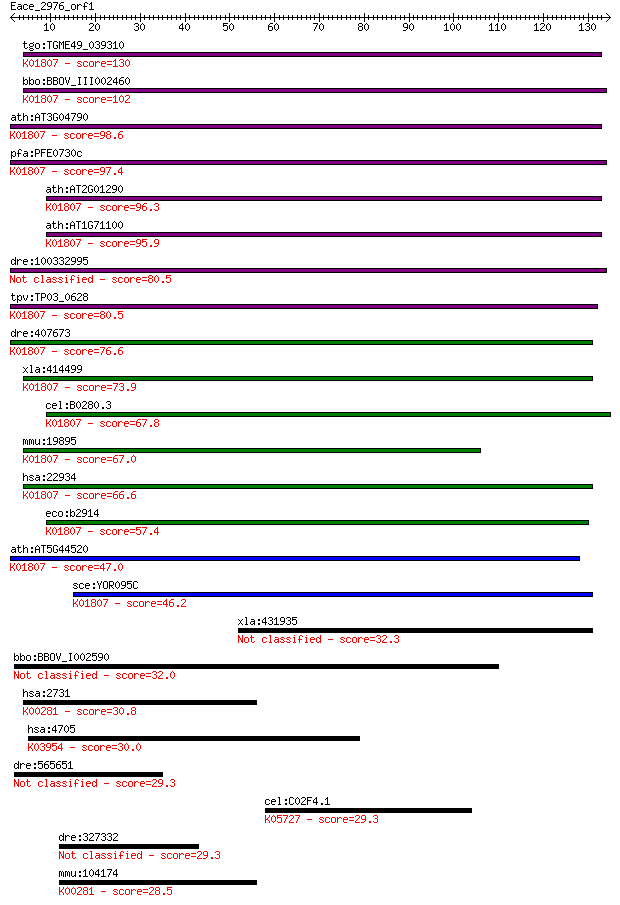
Query= Eace_2976_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_039310 ribose 5-phosphate isomerase, putative (EC:5... 130 1e-30
bbo:BBOV_III002460 17.m07239; ribose 5-phosphate isomerase A f... 102 3e-22
ath:AT3G04790 ribose 5-phosphate isomerase-related; K01807 rib... 98.6 5e-21
pfa:PFE0730c ribose 5-phosphate epimerase, putative (EC:5.3.1.... 97.4 1e-20
ath:AT2G01290 ribose-5-phosphate isomerase (EC:5.3.1.6); K0180... 96.3 2e-20
ath:AT1G71100 RSW10; RSW10 (RADIAL SWELLING 10); ribose-5-phos... 95.9 3e-20
dre:100332995 chloroplast ribose-5-phosphate isomerase-like 80.5 1e-15
tpv:TP03_0628 ribose 5-phosphate epimerase (EC:5.3.1.6); K0180... 80.5 1e-15
dre:407673 rpia, MGC103524, zgc:103524; ribose 5-phosphate iso... 76.6 2e-14
xla:414499 rpia, MGC83218; ribose 5-phosphate isomerase A (EC:... 73.9 1e-13
cel:B0280.3 hypothetical protein; K01807 ribose 5-phosphate is... 67.8 8e-12
mmu:19895 Rpia, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.... 67.0 2e-11
hsa:22934 RPIA, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.... 66.6 2e-11
eco:b2914 rpiA, ECK2910, JW5475, ygfC; ribose 5-phosphate isom... 57.4 1e-08
ath:AT5G44520 ribose 5-phosphate isomerase-related; K01807 rib... 47.0 1e-05
sce:YOR095C RKI1; Ribose-5-phosphate ketol-isomerase, catalyze... 46.2 2e-05
xla:431935 parp9, MGC83934, bal, bal1; poly (ADP-ribose) polym... 32.3 0.41
bbo:BBOV_I002590 19.m02862; hypothetical protein 32.0 0.48
hsa:2731 GLDC, GCE, GCSP, HYGN1, MGC138198, MGC138200; glycine... 30.8 1.3
hsa:4705 NDUFA10, CI-42KD, CI-42k, FLJ21923, MGC5103; NADH deh... 30.0 2.1
dre:565651 GDNF family receptor alpha 1 isoform a preproprotei... 29.3 3.4
cel:C02F4.1 ced-5; CEll Death abnormality family member (ced-5... 29.3 3.6
dre:327332 fi03c06; wu:fi03c06 29.3 3.8
mmu:104174 Gldc, D030049L12Rik, D19Wsu57e; glycine decarboxyla... 28.5 5.2
> tgo:TGME49_039310 ribose 5-phosphate isomerase, putative (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=259
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 75/137 (54%), Positives = 96/137 (70%), Gaps = 11/137 (8%)
Query 4 AVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVL 63
AV+T ++SGMKVGLGTG+TA F V R+ +R+Q+G L + C TSE TRK AESLGI +
Sbjct 15 AVDTYVRSGMKVGLGTGTTAKFVVERIGQRMQEGSLKDLLCVPTSEATRKQAESLGIPLT 74
Query 64 PLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE--------IVDEDKLVT 115
LD I+ LDVAIDGADE+ +L LVKGRGGAL+REK++ DE KLV+
Sbjct 75 TLDGIADCLDVAIDGADEILPP--TLGLVKGRGGALLREKMIAAAAKTFIVAADETKLVS 132
Query 116 DKTFGTTGALPLEIVQF 132
+ G+TGALP+E+V F
Sbjct 133 N-GIGSTGALPVEVVVF 148
> bbo:BBOV_III002460 17.m07239; ribose 5-phosphate isomerase A
family protein (EC:5.3.1.6); K01807 ribose 5-phosphate isomerase
A [EC:5.3.1.6]
Length=240
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 66/137 (48%), Positives = 87/137 (63%), Gaps = 11/137 (8%)
Query 4 AVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVL 63
AV+T IQ+GM VGLGTGSTAS+AVR LA+++ +L ++ +TS T +L LGI L
Sbjct 14 AVDTYIQNGMIVGLGTGSTASYAVRYLAKQLSAAKLRDVTAVATSVATFRLMSELGIPSL 73
Query 64 PLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVEI-VDEDKLVTDKTFGTT 122
D+ S+ +DVAIDGAD + L L+KG GGAL REKLVE+ + ++ D T
Sbjct 74 EFDD-SVSIDVAIDGADAI---DGDLNLIKGGGGALFREKLVELGARKVIIIADGTKKVK 129
Query 123 GAL------PLEIVQFG 133
G L P+EIVQFG
Sbjct 130 GHLLQAFHVPVEIVQFG 146
> ath:AT3G04790 ribose 5-phosphate isomerase-related; K01807 ribose
5-phosphate isomerase A [EC:5.3.1.6]
Length=276
Score = 98.6 bits (244), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 68/140 (48%), Positives = 90/140 (64%), Gaps = 13/140 (9%)
Query 1 ATEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGI 60
A E I+ GM +GLGTGSTA+FAV ++ + + GEL I TS+ T + A SLGI
Sbjct 52 AAEKAVEAIKPGMVLGLGTGSTAAFAVDQIGKLLSSGELYDIVGIPTSKRTEEQARSLGI 111
Query 61 SVLPLDEISLP-LDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVEIV-DEDKLVTDKT 118
++ LD + P +D+AIDGADEV +L LVKGRGGAL+REK+VE V D+ +V D T
Sbjct 112 PLVGLD--THPRIDLAIDGADEV---DPNLDLVKGRGGALLREKMVEAVADKFIVVADDT 166
Query 119 FGTTG------ALPLEIVQF 132
TG A+P+E+VQF
Sbjct 167 KLVTGLGGSGLAMPVEVVQF 186
> pfa:PFE0730c ribose 5-phosphate epimerase, putative (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=236
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 61/141 (43%), Positives = 86/141 (60%), Gaps = 13/141 (9%)
Query 1 ATEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGI 60
A +AV+ +QS M +GLGTGST + + R+ ++ G+L + C TS +T A LGI
Sbjct 9 AYKAVDEYVQSNMTIGLGTGSTVFYVLERIDNLLKSGKLKDVVCIPTSIDTELKARKLGI 68
Query 61 SVLPLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE--------IVDEDK 112
+ L++ S +D+ IDG DE+ +L L+KGRGGAL+REKLV I DE K
Sbjct 69 PLTTLEKHS-NIDITIDGTDEI---DLNLNLIKGRGGALVREKLVASSSSLFIIIGDESK 124
Query 113 LVTDKTFGTTGALPLEIVQFG 133
L T+ G TGA+P+EI+ FG
Sbjct 125 LCTNG-LGMTGAVPIEILTFG 144
> ath:AT2G01290 ribose-5-phosphate isomerase (EC:5.3.1.6); K01807
ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=265
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 63/132 (47%), Positives = 87/132 (65%), Gaps = 13/132 (9%)
Query 9 IQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVLPLDEI 68
++SGM +GLGTGSTA AV R+ E ++QG+L I TS++T++ A SLGI + LD
Sbjct 48 VESGMVLGLGTGSTAKHAVDRIGELLRQGKLENIVGIPTSKKTQEQALSLGIPLSDLDAH 107
Query 69 SLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE--------IVDEDKLVTDKTFG 120
+ +D++IDGADEV N LVKGRGG+L+REK++E IVD+ K+V G
Sbjct 108 PV-IDLSIDGADEVDPFLN---LVKGRGGSLLREKMIEGASKKFVVIVDDSKMVK-HIGG 162
Query 121 TTGALPLEIVQF 132
+ ALP+EIV F
Sbjct 163 SKLALPVEIVPF 174
> ath:AT1G71100 RSW10; RSW10 (RADIAL SWELLING 10); ribose-5-phosphate
isomerase (EC:5.3.1.6); K01807 ribose 5-phosphate isomerase
A [EC:5.3.1.6]
Length=267
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 64/133 (48%), Positives = 88/133 (66%), Gaps = 15/133 (11%)
Query 9 IQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVLPLDEI 68
++SGM +GLGTGSTA AV R++E +++G+L I TS T + A SLGI + LD
Sbjct 44 VESGMVIGLGTGSTAKHAVARISELLREGKLKDIIGIPTSTTTHEQAVSLGIPLSDLDSH 103
Query 69 SLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE--------IVDEDKLVTDKTFG 120
+ +D++IDGADEV +L LVKGRGG+L+REK++E IVDE KLV K G
Sbjct 104 PV-VDLSIDGADEV---DPALNLVKGRGGSLLREKMIEGASKKFVVIVDESKLV--KYIG 157
Query 121 TTG-ALPLEIVQF 132
+G A+P+E+V F
Sbjct 158 GSGLAVPVEVVPF 170
> dre:100332995 chloroplast ribose-5-phosphate isomerase-like
Length=291
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 60/138 (43%), Positives = 80/138 (57%), Gaps = 11/138 (7%)
Query 1 ATEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGI 60
A EA +++GM++G+GTGSTA VR LAE++ G + TSE T +L LG+
Sbjct 69 AAEAALQHVENGMRLGIGTGSTAEEFVRLLAEKVAGG--LKVEGVPTSERTARLCVELGV 126
Query 61 SVLPLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVEIVDEDKLVT---DK 117
+ LDE+ LD+ IDGADEV SL L+KG GGAL+REK+V + +V K
Sbjct 127 PLKSLDELP-ELDLTIDGADEV---DASLRLIKGGGGALLREKIVACASQRMIVIADETK 182
Query 118 TFGTTGA--LPLEIVQFG 133
GA LP+EI FG
Sbjct 183 VVDMLGAFPLPIEINAFG 200
> tpv:TP03_0628 ribose 5-phosphate epimerase (EC:5.3.1.6); K01807
ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=363
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 81/142 (57%), Gaps = 15/142 (10%)
Query 1 ATEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGI 60
A +AV+T ++S + LGTG T++FA+ RLA I+ +T I TS+ T A+ L I
Sbjct 14 AEKAVDTYVKSNSIIALGTGRTSTFALERLANHIKTKSITNILGIPTSQATYLKAKELEI 73
Query 61 SVLPLDEI---SLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE--------IVD 109
++ LDE+ +++AIDGAD V + L L+KG GGAL E +VE ++D
Sbjct 74 PLISLDEVIGSDRRINIAIDGADSV---DSDLNLIKGGGGALFTEFMVEQYSDQLIIMID 130
Query 110 EDKLVTDKTFGTTGALPLEIVQ 131
E KL D + LP+EIV+
Sbjct 131 ESKLCNDHLL-ESHYLPIEIVK 151
> dre:407673 rpia, MGC103524, zgc:103524; ribose 5-phosphate isomerase
A (ribose 5-phosphate epimerase) (EC:5.3.1.6); K01807
ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=275
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 55/138 (39%), Positives = 77/138 (55%), Gaps = 13/138 (9%)
Query 1 ATEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGI 60
A AV+ IQ+ +G+G+GST +AV RLAE+++Q +L I C TS + R+L G+
Sbjct 50 AYAAVDNHIQNNQVIGVGSGSTIVYAVDRLAEKVRQEKLN-IVCVPTSFQARQLILQHGL 108
Query 61 SVLPLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE------IVDEDKLV 114
+ LD LDVAIDGADEV +L L+KG GG L +EK+V IV D
Sbjct 109 PLSDLDR-HPELDVAIDGADEV---DTALTLIKGGGGCLTQEKIVAGCAKHFIVIADYRK 164
Query 115 TDKTFGTTG--ALPLEIV 130
K G +P+E++
Sbjct 165 DSKALGQQWKKGVPVEVI 182
> xla:414499 rpia, MGC83218; ribose 5-phosphate isomerase A (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=235
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 53/135 (39%), Positives = 75/135 (55%), Gaps = 13/135 (9%)
Query 4 AVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVL 63
AV+ +++ +G+G+GST AV RLAER++Q L + C TS + R+L G+ +
Sbjct 13 AVDNHVKNNQALGIGSGSTIVHAVNRLAERVKQENLK-VVCVPTSFQARQLILQNGLVLT 71
Query 64 PLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE------IVDEDKLVTDK 117
LD LDVAIDGADEV + L L+KG GG L +EK+V IV D K
Sbjct 72 DLDR-HPELDVAIDGADEV---DSDLNLIKGGGGCLAQEKIVAGCAKSFIVIADYRKDSK 127
Query 118 TFGTTG--ALPLEIV 130
FG +P+E++
Sbjct 128 NFGEQWKKGVPIEVI 142
> cel:B0280.3 hypothetical protein; K01807 ribose 5-phosphate
isomerase A [EC:5.3.1.6]
Length=251
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 50/134 (37%), Positives = 68/134 (50%), Gaps = 13/134 (9%)
Query 9 IQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVLPLDEI 68
+QSG ++G+G+GST + V L + Q G L I C TS T++ G+ V LD
Sbjct 30 VQSGCRLGVGSGSTVKYLVEYLKQGFQNGSLKDIICVPTSFLTKQWLIESGLPVSDLD-- 87
Query 69 SLP-LDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE------IVDEDKLVTDKTFGT 121
S P LDV IDGADEV +KG GG L +EK+V+ V D L K G
Sbjct 88 SHPELDVCIDGADEV---DGQFTCIKGGGGCLAQEKIVQTAAKNFYVIADYLKDSKHLGD 144
Query 122 -TGALPLEIVQFGA 134
+P+E++ A
Sbjct 145 RYPNVPIEVLPLAA 158
> mmu:19895 Rpia, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=303
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 43/102 (42%), Positives = 64/102 (62%), Gaps = 5/102 (4%)
Query 4 AVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVL 63
AV +++ +G+G+GST AV+R+AER++Q L I C TS + R+L G+++
Sbjct 82 AVENHVKNNQVLGIGSGSTIVHAVQRIAERVKQENLDLI-CIPTSFQARQLILQYGLTLS 140
Query 64 PLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLV 105
LD+ +D+AIDGADEV L L+KG GG L +EK+V
Sbjct 141 DLDQ-HPEIDLAIDGADEV---DAELNLIKGGGGCLTQEKIV 178
> hsa:22934 RPIA, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=311
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 74/135 (54%), Gaps = 13/135 (9%)
Query 4 AVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVL 63
AV +++ +G+G+GST AV+R+AER++Q L + C TS + R+L G+++
Sbjct 89 AVENHVRNNQVLGIGSGSTIVHAVQRIAERVKQENLN-LVCIPTSFQARQLILQYGLTLS 147
Query 64 PLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE------IVDEDKLVTDK 117
LD +D+AIDGADEV L L+KG GG L +EK+V IV D K
Sbjct 148 DLDR-HPEIDLAIDGADEV---DADLNLIKGGGGCLTQEKIVAGYASRFIVIADFRKDSK 203
Query 118 TFGTTG--ALPLEIV 130
G +P+E++
Sbjct 204 NLGDQWHKGIPIEVI 218
> eco:b2914 rpiA, ECK2910, JW5475, ygfC; ribose 5-phosphate isomerase,
constitutive (EC:5.3.1.6); K01807 ribose 5-phosphate
isomerase A [EC:5.3.1.6]
Length=219
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 67/124 (54%), Gaps = 11/124 (8%)
Query 9 IQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVLPLDEI 68
+Q G VG+GTGSTA+ + L +G++ G + +S+ T KL +SLGI V L+E+
Sbjct 18 VQPGTIVGVGTGSTAAHFIDALGT--MKGQIEG-AVSSSDASTEKL-KSLGIHVFDLNEV 73
Query 69 SLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVEIVDEDKLV---TDKTFGTTGAL 125
L + +DGADE+ + ++KG G AL REK++ V E + K G
Sbjct 74 D-SLGIYVDGADEI---NGHMQMIKGGGAALTREKIIASVAEKFICIADASKQVDILGKF 129
Query 126 PLEI 129
PL +
Sbjct 130 PLPV 133
> ath:AT5G44520 ribose 5-phosphate isomerase-related; K01807 ribose
5-phosphate isomerase A [EC:5.3.1.6]
Length=296
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 70/139 (50%), Gaps = 17/139 (12%)
Query 1 ATEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGI 60
A V+ ++SGM +GLG+G + FA+R L +++ G L + S + A GI
Sbjct 44 AHHTVDNYVKSGMIIGLGSGEASDFAIRYLGQQLGSGSLHNVVGVPMSARSASEAAKYGI 103
Query 61 SVLPLDEIS--LPLDVAIDGADEVFKTGNSLVLVKGRGGA-------LMREKLVEIVDED 111
PL+ + +D A AD V + N+L+ V GR + L ++ +V++ DE
Sbjct 104 ---PLEYYRDGVQIDFAFHDADAVEE--NTLIAVIGRRRSSQEDDYILKQKSIVKVADEA 158
Query 112 K-LVTDKTF--GTTGALPL 127
++ ++ + G G++P+
Sbjct 159 VFMIKEEQYKAGLEGSIPV 177
> sce:YOR095C RKI1; Ribose-5-phosphate ketol-isomerase, catalyzes
the interconversion of ribose 5-phosphate and ribulose 5-phosphate
in the pentose phosphate pathway; participates in
pyridoxine biosynthesis (EC:5.3.1.6); K01807 ribose 5-phosphate
isomerase A [EC:5.3.1.6]
Length=258
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/129 (27%), Positives = 63/129 (48%), Gaps = 17/129 (13%)
Query 15 VGLGTGSTASFAVRRLAERIQQGELTGIS----CASTSEETRKLAESLGISVLPLDEISL 70
+G+G+GST + R+ + + + ++ C T ++R L + + +++
Sbjct 42 IGIGSGSTVVYVAERIGQYLHDPKFYEVASKFICIPTGFQSRNLILDNKLQLGSIEQYPR 101
Query 71 PLDVAIDGADEVFKTGNSLVLVKGRGGALMREKLVE-------IVDEDKLVTDKTFGTTG 123
+D+A DGADEV +L L+KG G L +EKLV +V + + + K G
Sbjct 102 -IDIAFDGADEV---DENLQLIKGGGACLFQEKLVSTSAKTFIVVADSRKKSPKHLGKNW 157
Query 124 --ALPLEIV 130
+P+EIV
Sbjct 158 RQGVPIEIV 166
> xla:431935 parp9, MGC83934, bal, bal1; poly (ADP-ribose) polymerase
family, member 9 (EC:2.4.2.30)
Length=914
Score = 32.3 bits (72), Expect = 0.41, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 45/83 (54%), Gaps = 5/83 (6%)
Query 52 RKLAESLGISVLPLDEISLPLDVAIDGADEVFK--TGNSLVLVKGRGGALMREKLVEIVD 109
+KL+E L +SV D +D ++ A+E K G +L LVK GGA+++++ ++
Sbjct 74 KKLSEGLRVSVWKGDMTRQNVDAVVNAANEDLKHFGGLALALVKA-GGAVIQDESRRHIE 132
Query 110 EDKLVTDKTFGTT--GALPLEIV 130
+ K V + T G LP +++
Sbjct 133 KYKKVKSGSIAVTSAGNLPCKMI 155
> bbo:BBOV_I002590 19.m02862; hypothetical protein
Length=336
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 49/113 (43%), Gaps = 17/113 (15%)
Query 2 TEAVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGIS 61
TE NT ++S L T S S L E ++ E + S S E R + ES +
Sbjct 16 TEIRNTPLES-----LNTPSFRS----GLLEALEAAESSTDDSGSHSHELRPIVESYNVH 66
Query 62 VLPLDEISLPLDVAIDGADEVFKTGNSLVLV-----KGRGGALMREKLVEIVD 109
LPLD +SL + + F L LV K + AL+ E LV+ ++
Sbjct 67 FLPLDPVSL---RSTQASVYTFDGFPGLYLVRDFFTKEQCDALLLETLVDYIN 116
> hsa:2731 GLDC, GCE, GCSP, HYGN1, MGC138198, MGC138200; glycine
dehydrogenase (decarboxylating) (EC:1.4.4.2); K00281 glycine
dehydrogenase [EC:1.4.4.2]
Length=1020
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 4 AVNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLA 55
A+ + + G+ +G G A FAVR R+ G + G++ +T +E +LA
Sbjct 313 ALGSSQRFGVPLGYGGPHAAFFAVRESLVRMMPGRMVGVTRDATGKEVYRLA 364
> hsa:4705 NDUFA10, CI-42KD, CI-42k, FLJ21923, MGC5103; NADH dehydrogenase
(ubiquinone) 1 alpha subcomplex, 10, 42kDa (EC:1.6.5.3
1.6.99.3); K03954 NADH dehydrogenase (ubiquinone) 1
alpha subcomplex 10 [EC:1.6.5.3 1.6.99.3]
Length=355
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 32/74 (43%), Gaps = 0/74 (0%)
Query 5 VNTCIQSGMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLAESLGISVLP 64
+++ +Q ++ G+ A +RL ER + + G C + +++AE LG P
Sbjct 28 IHSSVQCKLRYGMWHFLLGDKASKRLTERSRVITVDGNICTGKGKLAKEIAEKLGFKHFP 87
Query 65 LDEISLPLDVAIDG 78
I P DG
Sbjct 88 EAGIHYPDSTTGDG 101
> dre:565651 GDNF family receptor alpha 1 isoform a preproprotein-like
Length=546
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 2 TEAVNTCIQSGMKVGLGTGSTASFAVRRLAERI 34
TE V+TCI+ K GL S + A+RR +R+
Sbjct 252 TEYVSTCIKPSTKSGLCNRSRCNKALRRFFDRV 284
> cel:C02F4.1 ced-5; CEll Death abnormality family member (ced-5);
K05727 dedicator of cytokinesis 3/4/5
Length=1781
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 58 LGISVLPLDEISLPLDVAIDGADEVFKTGNSLVLVKGRGGALMREK 103
LG V+ +E +PLDV + + +T + + KGR G+LMREK
Sbjct 155 LGQDVVIRNEEGVPLDVE---SLSLLRTYEAHISSKGRVGSLMREK 197
> dre:327332 fi03c06; wu:fi03c06
Length=514
Score = 29.3 bits (64), Expect = 3.8, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 12 GMKVGLGTGSTASFAVRRLAERIQQGELTGI 42
G+ VGLG G+ A A + A R GE T I
Sbjct 82 GLAVGLGIGAIAEVAKKSFASRDNSGENTSI 112
> mmu:104174 Gldc, D030049L12Rik, D19Wsu57e; glycine decarboxylase
(EC:1.4.4.2); K00281 glycine dehydrogenase [EC:1.4.4.2]
Length=1025
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 12 GMKVGLGTGSTASFAVRRLAERIQQGELTGISCASTSEETRKLA 55
G+ +G G A FAV+ R+ G + G++ +T +E +LA
Sbjct 326 GVPLGYGGPHAAFFAVKENLVRMMPGRMVGVTRDATGKEVYRLA 369
Lambda K H
0.314 0.133 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40