bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2993_orf5
Length=60
Score E
Sequences producing significant alignments: (Bits) Value
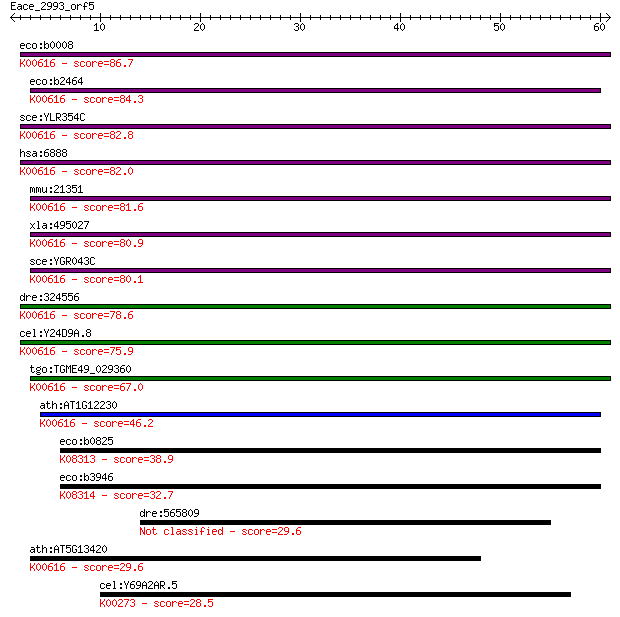
eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2... 86.7 2e-17
eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);... 84.3 8e-17
sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC... 82.8 2e-16
hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC... 82.0 4e-16
mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transal... 81.6 6e-16
xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transa... 80.9 1e-15
sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC... 80.1 2e-15
dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:6... 78.6 5e-15
cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.... 75.9 3e-14
tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616... 67.0 2e-11
ath:AT1G12230 transaldolase, putative; K00616 transaldolase [E... 46.2 3e-05
eco:b0825 fsaA, ECK0815, JW5109, mipB, ybiZ; fructose-6-phosph... 38.9 0.004
eco:b3946 fsaB, ECK3938, JW3918, talC, yijG; fructose-6-phosph... 32.7 0.33
dre:565809 fc15a01; wu:fc15a01 29.6 2.3
ath:AT5G13420 transaldolase, putative; K00616 transaldolase [E... 29.6 2.4
cel:Y69A2AR.5 hypothetical protein; K00273 D-amino-acid oxidas... 28.5 5.9
> eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=317
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/59 (64%), Positives = 52/59 (88%), Gaps = 0/59 (0%)
Query 2 TRERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
+ +RILIK+A+TW+GI+AA+QL++E INCN+TLLFS QA A ++AG +L+SPFVGRIL
Sbjct 125 SNDRILIKLASTWQGIRAAEQLEKEGINCNLTLLFSFAQARACAEAGVFLISPFVGRIL 183
> eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=316
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 37/57 (64%), Positives = 50/57 (87%), Gaps = 0/57 (0%)
Query 3 RERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRI 59
+ RILIK+A+TWEGI+AA++L++E INCN+TLLFS QA A ++AG +L+SPFVGRI
Sbjct 125 KSRILIKLASTWEGIRAAEELEKEGINCNLTLLFSFAQARACAEAGVFLISPFVGRI 181
> sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=335
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/60 (65%), Positives = 53/60 (88%), Gaps = 1/60 (1%)
Query 2 TRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
++ER+LIKIA+TWEGIQAAK+L+E++ I+CN+TLLFS QA+A ++A L+SPFVGRIL
Sbjct 137 SKERVLIKIASTWEGIQAAKELEEKDGIHCNLTLLFSFVQAVACAEAQVTLISPFVGRIL 196
> hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 37/60 (61%), Positives = 53/60 (88%), Gaps = 1/60 (1%)
Query 2 TRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
+++RILIK+++TWEGIQA K+L+E++ I+CN+TLLFS QA+A ++AG L+SPFVGRIL
Sbjct 135 SKDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFVGRIL 194
> mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=337
Score = 81.6 bits (200), Expect = 6e-16, Method: Composition-based stats.
Identities = 37/59 (62%), Positives = 52/59 (88%), Gaps = 1/59 (1%)
Query 3 RERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
++RILIK+++TWEGIQA K+L+E++ I+CN+TLLFS QA+A ++AG L+SPFVGRIL
Sbjct 136 KDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFVGRIL 194
> xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=338
Score = 80.9 bits (198), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/59 (64%), Positives = 50/59 (84%), Gaps = 1/59 (1%)
Query 3 RERILIKIAATWEGIQAAKQLKEE-NINCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
++RILIK+++TWEGIQA K L+EE I+CN+TLLFS QA+A ++AG L+SPFVGRIL
Sbjct 137 KKRILIKLSSTWEGIQAGKILEEEYGIHCNMTLLFSFAQAVACAEAGVTLISPFVGRIL 195
> sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=333
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/59 (61%), Positives = 50/59 (84%), Gaps = 1/59 (1%)
Query 3 RERILIKIAATWEGIQAAKQLK-EENINCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
+ER+LIKIA+TWEGIQAA++L+ + I+CN+TLLFS QA+A ++A L+SPFVGRI+
Sbjct 138 KERVLIKIASTWEGIQAARELEVKHGIHCNMTLLFSFTQAVACAEANVTLISPFVGRIM 196
> dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:66054;
transaldolase 1 (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 36/60 (60%), Positives = 50/60 (83%), Gaps = 1/60 (1%)
Query 2 TRERILIKIAATWEGIQAAKQLKE-ENINCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
++ERILIK+++TWEGIQA ++L+E I+CN+TLLFS QA+A ++A L+SPFVGRIL
Sbjct 135 SKERILIKLSSTWEGIQAGRELEEIHGIHCNMTLLFSFAQAVACAEARVTLISPFVGRIL 194
> cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.2.1.2]
Length=319
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/60 (56%), Positives = 52/60 (86%), Gaps = 1/60 (1%)
Query 2 TRERILIKIAATWEGIQAAKQLKEEN-INCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
+++RILIK+A+TWEGI+AAK L+ ++ I+CN+TLLF+ QA+A +++G L+SPFVGRI+
Sbjct 124 SKDRILIKLASTWEGIRAAKFLESKHGIHCNMTLLFNFEQAVACAESGVTLISPFVGRIM 183
> tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616
transaldolase [EC:2.2.1.2]
Length=364
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 37/58 (63%), Positives = 50/58 (86%), Gaps = 0/58 (0%)
Query 3 RERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRIL 60
++R+LIK+A TWEG +AAK L++E I+CN+TLLFS QA+AA++A A L+SPFVGRIL
Sbjct 161 KDRVLIKLATTWEGCEAAKILEKEGIHCNMTLLFSFAQAVAAAEAKATLISPFVGRIL 218
> ath:AT1G12230 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=405
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 28/56 (50%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 4 ERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRI 59
+R+L KI ATW+GI+AA+ L+ E I ++T ++S QA AAS AGA ++ FVGR+
Sbjct 196 DRLLFKIPATWQGIEAARLLESEGIQTHMTFVYSFAQAAAASQAGASVIQIFVGRL 251
> eco:b0825 fsaA, ECK0815, JW5109, mipB, ybiZ; fructose-6-phosphate
aldolase 1 (EC:4.1.2.-); K08313 fructose-6-phosphate aldolase
1 [EC:4.1.2.-]
Length=220
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 6 ILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRI 59
I++K+ T EG+ A K LK E I T ++ Q + ++ AGA V+P+V RI
Sbjct 82 IVVKVPVTAEGLAAIKMLKAEGIPTLGTAVYGAAQGLLSALAGAEYVAPYVNRI 135
> eco:b3946 fsaB, ECK3938, JW3918, talC, yijG; fructose-6-phosphate
aldolase 2 (EC:4.1.2.-); K08314 fructose-6-phosphate aldolase
2 [EC:4.1.2.-]
Length=220
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 6 ILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFVGRI 59
I++KI T EG+ A K LK+E I T ++S Q + A+ AGA V+P+V R+
Sbjct 82 IVVKIPVTSEGLAAIKILKKEGITTLGTAVYSAAQGLLAALAGAKYVAPYVNRV 135
> dre:565809 fc15a01; wu:fc15a01
Length=1345
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 11/41 (26%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 14 WEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSP 54
WEG+Q A+ + + + L + CQA+ + Y+ P
Sbjct 688 WEGVQEAEYWRRKIAMSPVMLSYGHCQAVPKQEQAVYVTDP 728
> ath:AT5G13420 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=438
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 3 RERILIKIAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDA 47
R + IKI AT I + + + I+ N+TL+FS+ + A DA
Sbjct 203 RRNVYIKIPATAPCIPSIRDVIAAGISVNVTLIFSIARYEAVIDA 247
> cel:Y69A2AR.5 hypothetical protein; K00273 D-amino-acid oxidase
[EC:1.4.3.3]
Length=322
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 10 IAATWEGIQAAKQLKEENINCNITLLFSLCQAIAASDAGAYLVSPFV 56
+ A GI +A ++E NC +T++ SD A L+ PF+
Sbjct 7 LGAGINGIASALAIQERLPNCEVTIIAEKFSPNTTSDVAAGLIEPFL 53
Lambda K H
0.322 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069361540
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40