bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3018_orf1
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
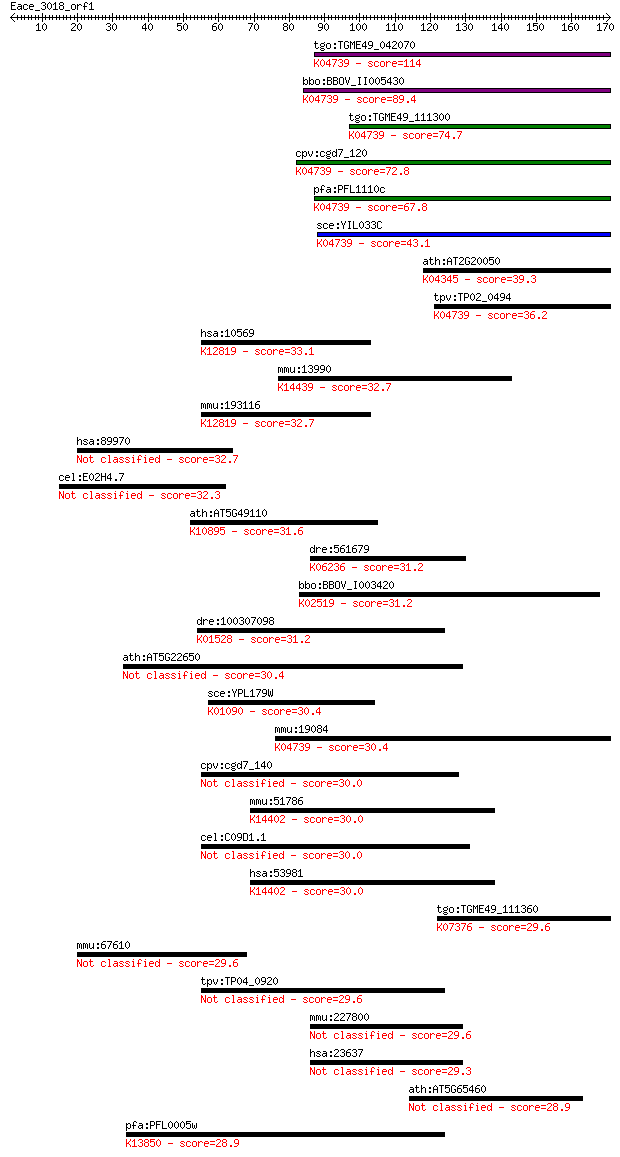
tgo:TGME49_042070 cAMP-dependent protein kinase regulatory sub... 114 2e-25
bbo:BBOV_II005430 18.m06452; cAMP-dependent protein kinase reg... 89.4 5e-18
tgo:TGME49_111300 cAMP-dependent protein kinase regulatory sub... 74.7 1e-13
cpv:cgd7_120 cAMP-dependent protein kinase regulatory subunit ... 72.8 5e-13
pfa:PFL1110c PKAr; CAMP-dependent protein kinase regulatory su... 67.8 2e-11
sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein k... 43.1 5e-04
ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regu... 39.3 0.006
tpv:TP02_0494 cAMP-dependent protein kinase regulatory subunit... 36.2 0.062
hsa:10569 SLU7, 9G8, MGC9280, hSlu7; SLU7 splicing factor homo... 33.1 0.52
mmu:13990 Smarcad1, AV081750, AW226546, D6Pas1, Etl1, mKIAA112... 32.7 0.55
mmu:193116 Slu7, AU018913, D11Ertd730e, D3Bwg0878e, MGC31026; ... 32.7 0.58
hsa:89970 RSPRY1, KIAA1972; ring finger and SPRY domain contai... 32.7 0.61
cel:E02H4.7 hypothetical protein 32.3 0.74
ath:AT5G49110 hypothetical protein; K10895 fanconi anemia grou... 31.6 1.2
dre:561679 col11a2, col11a1; collagen type XI alpha-2; K06236 ... 31.2 1.7
bbo:BBOV_I003420 19.m02088; elongation factor Tu GTP binding d... 31.2 1.9
dre:100307098 dnm1; si:dkey-246l19.2; K01528 dynamin GTPase [E... 31.2 1.9
ath:AT5G22650 HD2B; HD2B (HISTONE DEACETYLASE 2B); histone dea... 30.4 3.3
sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein ph... 30.4 3.4
mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protei... 30.4 3.4
cpv:cgd7_140 SprT family metallopeptidase 30.0 3.5
mmu:51786 Cpsf2, 100kDa, 2610024B04Rik, AI662483, Cpsf, MCPSF,... 30.0 3.9
cel:C09D1.1 unc-89; UNCoordinated family member (unc-89) 30.0 4.1
hsa:53981 CPSF2, CPSF100, KIAA1367; cleavage and polyadenylati... 30.0 4.5
tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:... 29.6 4.7
mmu:67610 Rspry1, 4930470D19Rik, AI608258; ring finger and SPR... 29.6 4.7
tpv:TP04_0920 hypothetical protein 29.6 5.0
mmu:227800 Rabgap1, Gapcena, KIAA4104, MGC30493, mKIAA4104; RA... 29.6 5.1
hsa:23637 RABGAP1, DKFZp586D2123, GAPCENA, RP11-123N4.2, TBC1D... 29.3 6.2
ath:AT5G65460 kinesin motor protein-related 28.9 9.1
pfa:PFL0005w VAR; erythrocyte membrane protein 1, PfEMP1; K138... 28.9 9.9
> tgo:TGME49_042070 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.10.2); K04739 cAMP-dependent protein
kinase regulator
Length=308
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/86 (62%), Positives = 70/86 (81%), Gaps = 2/86 (2%)
Query 87 LPMPSAK--AQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILENSFLFSSL 144
+P PS ++AKNRQSVSAE YGEWNK+K FVAPV+ KT E+K+RI ++E+SFLFSSL
Sbjct 1 MPQPSEAYMSRAKNRQSVSAEAYGEWNKRKKFVAPVYPKTAEQKERITKVIESSFLFSSL 60
Query 145 EAEDIEVVLGAFEGLSVAKGTVIIKQ 170
+ ED+E V+ AF+ +SV KGTVII+Q
Sbjct 61 DIEDLETVINAFQEVSVKKGTVIIRQ 86
> bbo:BBOV_II005430 18.m06452; cAMP-dependent protein kinase regulatory
subunit; K04739 cAMP-dependent protein kinase regulator
Length=317
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 44/91 (48%), Positives = 64/91 (70%), Gaps = 4/91 (4%)
Query 84 VDDLPMPSAKA----QAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILENSF 139
DD+ SA+A +A+NR S+SAEV+G +N FVAPVHEKTPE+ RI++ + F
Sbjct 3 TDDIDAISAEAIRFARARNRVSISAEVFGAYNNPDLFVAPVHEKTPEQATRIKETVLKCF 62
Query 140 LFSSLEAEDIEVVLGAFEGLSVAKGTVIIKQ 170
LFSS++A+DI+ ++ AFE V+ GT +I+Q
Sbjct 63 LFSSVDAKDIDTLVKAFESSDVSAGTKVIQQ 93
> tgo:TGME49_111300 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.11.12); K04739 cAMP-dependent protein
kinase regulator
Length=410
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/74 (50%), Positives = 52/74 (70%), Gaps = 0/74 (0%)
Query 97 KNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILENSFLFSSLEAEDIEVVLGAF 156
K R SVSAEVYGEWNKKK FVAPV+EK +K+++ IL SFLF+SL+ +D+ V+ A
Sbjct 122 KMRCSVSAEVYGEWNKKKNFVAPVYEKDEGQKEQLERILRQSFLFNSLDEKDLNTVILAM 181
Query 157 EGLSVAKGTVIIKQ 170
+ + T +I++
Sbjct 182 QEKKIEASTCLIRE 195
> cpv:cgd7_120 cAMP-dependent protein kinase regulatory subunit
; K04739 cAMP-dependent protein kinase regulator
Length=345
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/89 (44%), Positives = 55/89 (61%), Gaps = 0/89 (0%)
Query 82 DVVDDLPMPSAKAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILENSFLF 141
DV D++ +P R SVSAE YG WNK K F P + KT E++KRIR+ L SF+F
Sbjct 41 DVRDNIEIPKNFLARGPRTSVSAEAYGAWNKMKDFTPPSYPKTKEQEKRIREKLLESFMF 100
Query 142 SSLEAEDIEVVLGAFEGLSVAKGTVIIKQ 170
+SL+ ++++ V+ A SV K T II Q
Sbjct 101 TSLDDDELKTVILACVETSVKKDTEIITQ 129
> pfa:PFL1110c PKAr; CAMP-dependent protein kinase regulatory
subunit, putative; K04739 cAMP-dependent protein kinase regulator
Length=441
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/85 (42%), Positives = 52/85 (61%), Gaps = 1/85 (1%)
Query 87 LPMPSAKAQAKNRQSVSAEVYGEWNKK-KPFVAPVHEKTPEEKKRIRDILENSFLFSSLE 145
L + S K R SVSAE YG+WNKK F+ V++K +EK +IR+ L SFLF+ L
Sbjct 135 LDLESIHFIQKKRLSVSAEAYGDWNKKIDNFIPKVYKKDEKEKAKIREALNESFLFNHLN 194
Query 146 AEDIEVVLGAFEGLSVAKGTVIIKQ 170
++ E+++ AF +V KG II +
Sbjct 195 KKEFEIIVNAFFDKNVEKGVNIINE 219
> sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein
kinase regulator
Length=416
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 46/90 (51%), Gaps = 15/90 (16%)
Query 88 PMPSAKAQAKNRQSVSAEV-----YGEWNKKKPFVAPVH--EKTPEEKKRIRDILENSFL 140
P+P A+ R SVS E + +W P H EK+ ++ +R+ + N+FL
Sbjct 133 PLP-MHFNAQRRTSVSGETLQPNNFDDW-------TPDHYKEKSEQQLQRLEKSIRNNFL 184
Query 141 FSSLEAEDIEVVLGAFEGLSVAKGTVIIKQ 170
F+ L+++ +V+ E SV KG IIKQ
Sbjct 185 FNKLDSDSKRLVINCLEEKSVPKGATIIKQ 214
> ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regulator/
catalytic/ protein kinase/ protein serine/threonine
phosphatase; K04345 protein kinase A [EC:2.7.11.11]
Length=1094
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 118 APVHEKTPEEKKRIRDILENSFLFSSLEAEDIEVVLGAFEGLSVAKGTVIIKQ 170
+P H KT EE+ I +L + FLF L +V+L + L G +++KQ
Sbjct 469 SPAHRKTWEEEAHIERVLRDHFLFRKLTDSQCQVLLDCMQRLEANPGDIVVKQ 521
> tpv:TP02_0494 cAMP-dependent protein kinase regulatory subunit;
K04739 cAMP-dependent protein kinase regulator
Length=285
Score = 36.2 bits (82), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 37/56 (66%), Gaps = 6/56 (10%)
Query 121 HEKTPEEKKRIRDILENSFLFSSLEAEDIEVVLGAFE------GLSVAKGTVIIKQ 170
+EKTP+E+ +I +++++ FLFS + + +++++ AF+ L + G V+IKQ
Sbjct 13 YEKTPQEQAKIMEMIKSCFLFSGVTSSGLDLLVKAFDFKAAKIKLFSSAGDVLIKQ 68
> hsa:10569 SLU7, 9G8, MGC9280, hSlu7; SLU7 splicing factor homolog
(S. cerevisiae); K12819 pre-mRNA-processing factor SLU7
Length=586
Score = 33.1 bits (74), Expect = 0.52, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 55 DSPNSQAERERPPQQVTEDDLDSNEDDD-VVDDLPMPSAKAQAKNRQSV 102
+SP Q E P Q+ +D +ED+D DD+ MP +K R +V
Sbjct 214 NSPKHQWGEEEPNSQMEKDHNSEDEDEDKYADDIDMPGQNFDSKRRITV 262
> mmu:13990 Smarcad1, AV081750, AW226546, D6Pas1, Etl1, mKIAA1122;
SWI/SNF-related, matrix-associated actin-dependent regulator
of chromatin, subfamily a, containing DEAD/H box 1 (EC:3.6.4.12);
K14439 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A containing DEAD/H
box 1 [EC:3.6.4.12]
Length=1021
Score = 32.7 bits (73), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query 77 SNEDDDVVDDLPMPSAKAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPE-EKKRIRDIL 135
S+E+DDV DD + + + SAE W K++ V + ++ P +K+ +R++L
Sbjct 212 SSEEDDVNDDQSVKQPRGDRGEESNESAEASSNWEKQESIVLKLQKEFPNFDKQELREVL 271
Query 136 -ENSFLFS 142
E+ ++++
Sbjct 272 KEHEWMYT 279
> mmu:193116 Slu7, AU018913, D11Ertd730e, D3Bwg0878e, MGC31026;
SLU7 splicing factor homolog (S. cerevisiae); K12819 pre-mRNA-processing
factor SLU7
Length=585
Score = 32.7 bits (73), Expect = 0.58, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 55 DSPNSQAERERPPQQVTEDDLDSNEDDD-VVDDLPMPSAKAQAKNRQSV 102
+SP Q E P Q+ +D +ED+D DD+ MP +K R +V
Sbjct 214 NSPKHQWGEEEPNSQMEKDHNSEDEDEDKYADDIDMPGQNFDSKRRITV 262
> hsa:89970 RSPRY1, KIAA1972; ring finger and SPRY domain containing
1
Length=576
Score = 32.7 bits (73), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 20 LLLLLLYQLAGFAFSLSRVSTMGNVCECRGKVGGRDSPNSQAER 63
LLL L +A F + +TMGN C CR G DS ++Q ++
Sbjct 19 LLLTLEEHIAHFLGTGGAATTMGNSCICRDDSGTDDSVDTQQQQ 62
> cel:E02H4.7 hypothetical protein
Length=150
Score = 32.3 bits (72), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 25/47 (53%), Gaps = 5/47 (10%)
Query 15 LLLLLLLLLLLYQLAGFAFSLSRVSTMGNVCECRGKVGGRDSPNSQA 61
+L +L +LLL F F+ S VST G VC G GR +P S +
Sbjct 1 MLKWMLTVLLL-----FTFAFSGVSTTGCVCYEAGNYTGRSAPISNS 42
> ath:AT5G49110 hypothetical protein; K10895 fanconi anemia group
I protein
Length=1470
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 25/53 (47%), Gaps = 6/53 (11%)
Query 52 GGRDSPNSQAERERPPQQVTEDDLDSNEDDDVVDDLPMPSAKAQAKNRQSVSA 104
GG D N + E E + EDDL DDD +++ P +N SVS+
Sbjct 1369 GGEDGGNQEEETETQNNNIGEDDLRQESDDDGSEEMLSP------RNASSVSS 1415
> dre:561679 col11a2, col11a1; collagen type XI alpha-2; K06236
collagen, type I/II/III/V/XI, alpha
Length=1695
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 86 DLPMPSAKAQAKN--RQSVSAEVYGEWNKKKPFVAPVHEKTPEEKK 129
D P+P ++Q N R+S + K+KP P ++KTP+ K+
Sbjct 233 DSPLPKVQSQDPNTYRRSKHNQPKARQTKRKPMRPPQNQKTPKPKQ 278
> bbo:BBOV_I003420 19.m02088; elongation factor Tu GTP binding
domain containing protein; K02519 translation initiation factor
IF-2
Length=893
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 45/85 (52%), Gaps = 2/85 (2%)
Query 83 VVDDLPMPSAKAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILENSFLFS 142
+VD+L +P + + A+ + SV + + +P V + K + D L+NS + S
Sbjct 333 IVDELSLPYSVSIAEYQGSVLLKRIKQSMVTRPLVVTIMGHVDHGKTTLLDTLQNSDIVS 392
Query 143 SLEAEDIEVVLGAFEGLSVAKGTVI 167
EA I LGAF+ L++ +GT++
Sbjct 393 G-EAGRITQKLGAFK-LNLDRGTLV 415
> dre:100307098 dnm1; si:dkey-246l19.2; K01528 dynamin GTPase
[EC:3.6.5.5]
Length=1210
Score = 31.2 bits (69), Expect = 1.9, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 31/70 (44%), Gaps = 7/70 (10%)
Query 54 RDSPNSQAERERPPQQVTEDDLDSNEDDDVVDDLPMPSAKAQAKNRQSVSAEVYGEWNKK 113
RDS ++ PP E+ +++ D D P+ A A A + SVSA K
Sbjct 242 RDSLPEESVVNEPPATEAEEQCQADKTDAADVDQPISGAPADAGSSDSVSA-------VK 294
Query 114 KPFVAPVHEK 123
+P APV +
Sbjct 295 QPEAAPVQHQ 304
> ath:AT5G22650 HD2B; HD2B (HISTONE DEACETYLASE 2B); histone deacetylase
Length=305
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 40/96 (41%), Gaps = 19/96 (19%)
Query 33 FSLSRVSTMGNVCECRGKVGGRDSPNSQAERERPPQQVTEDDLDSNEDDDVVDDLPMPSA 92
F LS T NV G SPN + +DD S++D+DV + +P P+
Sbjct 75 FELSHSGTKANVHFI-----GYKSPN-----------IEQDDFTSSDDEDVPEAVPAPAP 118
Query 93 KAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEK 128
A N + +A V + KP P K EEK
Sbjct 119 TAVTANGNAGAAVVKADT---KPKAKPAEVKPAEEK 151
> sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=549
Score = 30.4 bits (67), Expect = 3.4, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 57 PNSQAERERPPQQVT--EDDLDSNEDDDVVDDLPMPSAKAQAKNRQSVS 103
PN QQ+T DDL+SNE +D D +P+ K + N+ S++
Sbjct 15 PNCSTNSNSSQQQLTTPSDDLNSNEPNDPDDSRSLPTIK-KFNNKHSIN 62
> mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protein
kinase, cAMP dependent regulatory, type I, alpha (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 30.4 bits (67), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 45/95 (47%), Gaps = 3/95 (3%)
Query 76 DSNEDDDVVDDLPMPSAKAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDIL 135
DS ED+ + P P K + + R ++SAEVY E + +V V K + + +
Sbjct 76 DSREDE-ISPPPPNPVVKGR-RRRGAISAEVYTEEDAAS-YVRKVIPKDYKTMAALAKAI 132
Query 136 ENSFLFSSLEAEDIEVVLGAFEGLSVAKGTVIIKQ 170
E + LFS L+ + + A +S G +I+Q
Sbjct 133 EKNVLFSHLDDNERSDIFDAMFPVSFIAGETVIQQ 167
> cpv:cgd7_140 SprT family metallopeptidase
Length=598
Score = 30.0 bits (66), Expect = 3.5, Method: Composition-based stats.
Identities = 18/77 (23%), Positives = 39/77 (50%), Gaps = 9/77 (11%)
Query 55 DSPNSQAERERPPQQVTEDDLDSNEDDDVVDDLPMPSAK----AQAKNRQSVSAEVYGEW 110
D + +++ E + V+ DL+SN+ ++ ++P+ + A + R S++ Y ++
Sbjct 308 DIEDQKSDLEEIKENVSSKDLNSNQVTNLTKEIPLKNKSEKFGAFIRQRSSIAKYWYQKF 367
Query 111 NKKKPFVAPVHEKTPEE 127
NK+ H + PEE
Sbjct 368 NKQV-----FHNRLPEE 379
> mmu:51786 Cpsf2, 100kDa, 2610024B04Rik, AI662483, Cpsf, MCPSF,
mKIAA1367; cleavage and polyadenylation specific factor 2;
K14402 cleavage and polyadenylation specificity factor subunit
2
Length=782
Score = 30.0 bits (66), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 40/96 (41%), Gaps = 27/96 (28%)
Query 69 QVTEDDLDSNEDDDVVDDLPMPSAK-------------------AQAKN--------RQS 101
Q E D+DS+++ DV +D+ PSA QAK +
Sbjct 411 QSKEADIDSSDESDVEEDVDQPSAHKTKHDLMMKGEGSRKGSFFKQAKKSYPMFPAPEER 470
Query 102 VSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILEN 137
+ + YGE K + F+ P + T EEK ++ L N
Sbjct 471 IKWDEYGEIIKPEDFLVPELQATEEEKSKLESGLTN 506
> cel:C09D1.1 unc-89; UNCoordinated family member (unc-89)
Length=8081
Score = 30.0 bits (66), Expect = 4.1, Method: Composition-based stats.
Identities = 20/80 (25%), Positives = 37/80 (46%), Gaps = 5/80 (6%)
Query 55 DSPNSQAERERPPQQVTEDDLDS----NEDDDVVDDLPMPSAKAQAKNRQSVSAEVYGEW 110
+ P S ++E P + T+D++ S + V++ P K + +SV EV
Sbjct 1664 EKPTSPTKKESSPTKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEKSPEKSVVEEVKSP- 1722
Query 111 NKKKPFVAPVHEKTPEEKKR 130
+K P A K+P +K++
Sbjct 1723 KEKSPEKAEEKPKSPTKKEK 1742
> hsa:53981 CPSF2, CPSF100, KIAA1367; cleavage and polyadenylation
specific factor 2, 100kDa; K14402 cleavage and polyadenylation
specificity factor subunit 2
Length=782
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 40/96 (41%), Gaps = 27/96 (28%)
Query 69 QVTEDDLDSNEDDDVVDDLPMPSAK-------------------AQAKN--------RQS 101
Q E D+DS+++ D+ +D+ PSA QAK +
Sbjct 411 QSKEADIDSSDESDIEEDIDQPSAHKTKHDLMMKGEGSRKGSFFKQAKKSYPMFPAPEER 470
Query 102 VSAEVYGEWNKKKPFVAPVHEKTPEEKKRIRDILEN 137
+ + YGE K + F+ P + T EEK ++ L N
Sbjct 471 IKWDEYGEIIKPEDFLVPELQATEEEKSKLESGLTN 506
> tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=994
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 122 EKTPEEKKRIRDILENSFLFSSLEAEDIEVVLGAFEGLSVAKGTVIIKQ 170
EKTP + I+D L+ + + SSL +I+ + A + + KG V+ KQ
Sbjct 171 EKTPADFALIQDSLKANLVCSSLNEGEIDALAVAMQFFTFKKGDVVTKQ 219
> mmu:67610 Rspry1, 4930470D19Rik, AI608258; ring finger and SPRY
domain containing 1
Length=576
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 3/51 (5%)
Query 20 LLLLLLYQLAGFAFSLSRVSTMGNVCECRGKVGGRDSPNS---QAERERPP 67
LLL L +A + +TMGN C CR G D+ ++ QAE P
Sbjct 19 LLLTLEEHIAHLLGTTGATATMGNSCICRDDSGAEDNVDTHQQQAENSTVP 69
> tpv:TP04_0920 hypothetical protein
Length=547
Score = 29.6 bits (65), Expect = 5.0, Method: Composition-based stats.
Identities = 19/79 (24%), Positives = 35/79 (44%), Gaps = 10/79 (12%)
Query 55 DSPNSQ-AERERPPQQVTEDDL---------DSNEDDDVVDDLPMPSAKAQAKNRQSVSA 104
D+PN A E + V+ D+ +S++DD D +P S + + A
Sbjct 22 DNPNDDLANTEDTIENVSGDEFVNFLVSEATESSKDDQKQDSIPQTSGEVGIEEETQEEA 81
Query 105 EVYGEWNKKKPFVAPVHEK 123
+VY E +K + P+ ++
Sbjct 82 KVYAELQPQKLYQEPIKQE 100
> mmu:227800 Rabgap1, Gapcena, KIAA4104, MGC30493, mKIAA4104;
RAB GTPase activating protein 1
Length=809
Score = 29.6 bits (65), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 86 DLPMPSAKAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEK 128
D+ + ++ K+ S + G WN K P V+E+TP++K
Sbjct 356 DMHLLDLESMGKSSDGKSYVITGSWNPKSPHFQVVNEETPKDK 398
> hsa:23637 RABGAP1, DKFZp586D2123, GAPCENA, RP11-123N4.2, TBC1D11;
RAB GTPase activating protein 1
Length=1069
Score = 29.3 bits (64), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 86 DLPMPSAKAQAKNRQSVSAEVYGEWNKKKPFVAPVHEKTPEEK 128
D+ + ++ K+ S + G WN K P V+E+TP++K
Sbjct 361 DMHLLDLESMGKSSDGKSYVITGSWNPKSPHFQVVNEETPKDK 403
> ath:AT5G65460 kinesin motor protein-related
Length=1281
Score = 28.9 bits (63), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 114 KPFVA-PVHEKTPEEKKRIRDILENSFLFSSLEAEDIE-VVLGAFEGLSVA 162
KP A VH E +++IR L SF F S+ A+D+ V G E LS A
Sbjct 908 KPLAALFVHTPAGELQRQIRSWLAESFEFLSVTADDVSGVTTGQLELLSTA 958
> pfa:PFL0005w VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2178
Score = 28.9 bits (63), Expect = 9.9, Method: Composition-based stats.
Identities = 19/97 (19%), Positives = 44/97 (45%), Gaps = 7/97 (7%)
Query 34 SLSRVSTMGNVCECRGKVGGRDSPNSQAERERPPQQVTEDDLDSNEDDDVVDDLPMPSAK 93
S + + GN GK D+ N + P ++T+++ ++ +D+ + + L K
Sbjct 1792 SGNNTTASGNNTTASGKNTPSDTQNDIPSGDTPNNKLTDNEWNTLKDEFISNMLQSEQPK 1851
Query 94 AQAKNRQS-------VSAEVYGEWNKKKPFVAPVHEK 123
+ S + +Y + N++KPF+ +H++
Sbjct 1852 DVPNDYSSGDIPFNTQPSTLYFDNNQEKPFITSIHDR 1888
Lambda K H
0.318 0.137 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4287611064
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40