bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3056_orf3
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
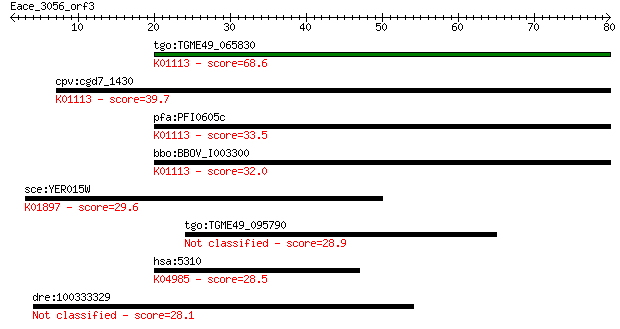
tgo:TGME49_065830 hypothetical protein ; K01113 alkaline phosp... 68.6 5e-12
cpv:cgd7_1430 possible phosphodiesterase/alkaline phosphatase ... 39.7 0.003
pfa:PFI0605c conserved Plasmodium protein, unknown function; K... 33.5 0.15
bbo:BBOV_I003300 19.m02166; hypothetical protein; K01113 alkal... 32.0 0.55
sce:YER015W FAA2, FAM1; Faa2p (EC:6.2.1.3); K01897 long-chain ... 29.6 2.8
tgo:TGME49_095790 hypothetical protein 28.9 4.2
hsa:5310 PKD1, PBP, Pc-1, TRPP1; polycystic kidney disease 1 (... 28.5 4.9
dre:100333329 MAP7 domain containing 2-like 28.1 8.0
> tgo:TGME49_065830 hypothetical protein ; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=614
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 41/60 (68%), Gaps = 0/60 (0%)
Query 20 VFETVAAKKPDGWIWIGDAGYAEGPDVASVEASLSAVSATQSYKTLKANISFLDGTWDDN 79
V+ V + P+ WIW+GDAGYA+ DVA+V +L V ++ L+A++ FLDGTWDD+
Sbjct 196 VWHRVKEQSPNAWIWMGDAGYAQSHDVAAVRLALKQVKDVDPFRELRASLHFLDGTWDDH 255
> cpv:cgd7_1430 possible phosphodiesterase/alkaline phosphatase
D, of possible plant or bacterial origin ; K01113 alkaline
phosphatase D [EC:3.1.3.1]
Length=463
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 35/74 (47%), Gaps = 1/74 (1%)
Query 7 GQRQIAAAAR-REKVFETVAAKKPDGWIWIGDAGYAEGPDVASVEASLSAVSATQSYKTL 65
GQ+Q + ++ ++ + + KPD W+GD+ Y + V S Y+ L
Sbjct 39 GQKQDKSGNYVSQEYWKAIQSDKPDALFWMGDSVYTKCGSPVCVSKGYLVQSNNSFYRDL 98
Query 66 KANISFLDGTWDDN 79
F+DGTWDD+
Sbjct 99 LKTGLFVDGTWDDH 112
> pfa:PFI0605c conserved Plasmodium protein, unknown function;
K01113 alkaline phosphatase D [EC:3.1.3.1]
Length=446
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 20 VFETVAAKKPDGWIWIGDAGYAEGPDVASVEASLSAVSATQSYKTLKANISFLDGTWDDN 79
+ ++ +KP +WIGD Y E ++ ++ + + + Y LK +DG +DD+
Sbjct 49 LLNSIEKRKPQLMLWIGDYFYTECSEIKCLDDAYTYIKKDPFYMKLKKKFK-IDGIYDDH 107
> bbo:BBOV_I003300 19.m02166; hypothetical protein; K01113 alkaline
phosphatase D [EC:3.1.3.1]
Length=754
Score = 32.0 bits (71), Expect = 0.55, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 32/64 (50%), Gaps = 6/64 (9%)
Query 20 VFETVAAKKPDGWIWIGDAGYAE----GPDVASVEASLSAVSATQSYKTLKANISFLDGT 75
+F+ + PD +++ GD Y E P +++ + + Y+ KANI +DG
Sbjct 66 IFQRIIDSNPDLFLYTGDIVYPEYGCCSPKC--LQSKYEQLVSHPVYQHFKANIRRIDGV 123
Query 76 WDDN 79
+DD+
Sbjct 124 YDDH 127
> sce:YER015W FAA2, FAM1; Faa2p (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=744
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 23/51 (45%), Gaps = 4/51 (7%)
Query 3 KESKGQRQIAAAARREKVF----ETVAAKKPDGWIWIGDAGYAEGPDVASV 49
K+ KG+ QI E+ F ET A DGW GD + +G SV
Sbjct 536 KDLKGELQIRGPQVFERYFKNPNETSKAVDQDGWFSTGDVAFIDGKGRISV 586
> tgo:TGME49_095790 hypothetical protein
Length=1386
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Query 24 VAAKKPD--GWIWIGDAGYAEGPDVASVEASLSAVSATQSYKT 64
+ K+P+ GW AG+ PDVAS+ +LS +S +T
Sbjct 96 LPEKRPEQNGWQLAAGAGHQSRPDVASLTHALSGSQTEESPRT 138
> hsa:5310 PKD1, PBP, Pc-1, TRPP1; polycystic kidney disease 1
(autosomal dominant); K04985 polycystin 1
Length=4302
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 20 VFETVAAKKPDGWIW-IGDAGYAEGPDV 46
+F V +P ++W +GD G+ EGP+V
Sbjct 1486 LFSAVGRGRPASYLWDLGDGGWLEGPEV 1513
> dre:100333329 MAP7 domain containing 2-like
Length=772
Score = 28.1 bits (61), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 21/55 (38%), Gaps = 5/55 (9%)
Query 4 ESKGQRQIAAAARREKVFETVAAKKPDGWIWIGDAGYAEG-----PDVASVEASL 53
E Q ++ A RR A ++P W W G G EG P ASL
Sbjct 206 ERNEQERLEALMRRSTNRNLQADQRPKRWTWCGPPGACEGDSKIAPPCPPASASL 260
Lambda K H
0.311 0.125 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2063098576
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40