bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3057_orf2
Length=74
Score E
Sequences producing significant alignments: (Bits) Value
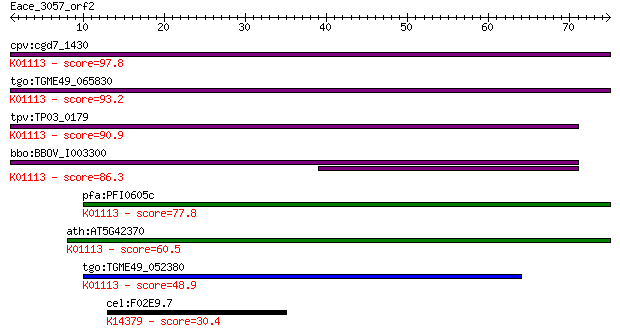
cpv:cgd7_1430 possible phosphodiesterase/alkaline phosphatase ... 97.8 8e-21
tgo:TGME49_065830 hypothetical protein ; K01113 alkaline phosp... 93.2 2e-19
tpv:TP03_0179 hypothetical protein; K01113 alkaline phosphatas... 90.9 9e-19
bbo:BBOV_I003300 19.m02166; hypothetical protein; K01113 alkal... 86.3 2e-17
pfa:PFI0605c conserved Plasmodium protein, unknown function; K... 77.8 7e-15
ath:AT5G42370 hypothetical protein; K01113 alkaline phosphatas... 60.5 1e-09
tgo:TGME49_052380 hypothetical protein ; K01113 alkaline phosp... 48.9 4e-06
cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant ac... 30.4 1.6
> cpv:cgd7_1430 possible phosphodiesterase/alkaline phosphatase
D, of possible plant or bacterial origin ; K01113 alkaline
phosphatase D [EC:3.1.3.1]
Length=463
Score = 97.8 bits (242), Expect = 8e-21, Method: Composition-based stats.
Identities = 42/74 (56%), Positives = 54/74 (72%), Gaps = 0/74 (0%)
Query 1 LGVGYEYEADVLGEEQWGWLEAQLKHSRAQAHIVVSSIQVLTGLPLVESWGLFPVSRLRL 60
G G+ Y+ D+LG+EQW WLE +L +S+A A+I++SSIQV T P+VE WG FP SR RL
Sbjct 211 FGYGHSYQGDMLGKEQWDWLEKELSNSKALANIIISSIQVTTQYPIVEGWGHFPKSRERL 270
Query 61 LSLFARTKPKGLML 74
SL +TKPKGL
Sbjct 271 FSLIKKTKPKGLFF 284
> tgo:TGME49_065830 hypothetical protein ; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=614
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 43/74 (58%), Positives = 53/74 (71%), Gaps = 0/74 (0%)
Query 1 LGVGYEYEADVLGEEQWGWLEAQLKHSRAQAHIVVSSIQVLTGLPLVESWGLFPVSRLRL 60
+G YE D+LGEEQW WLEAQL +S A H++VSSIQV T LPLVESWG FP ++ RL
Sbjct 445 FSLGGNYEGDILGEEQWRWLEAQLTNSTASVHLIVSSIQVSTTLPLVESWGHFPRAKQRL 504
Query 61 LSLFARTKPKGLML 74
+ L TKP G++
Sbjct 505 IDLLESTKPAGVLF 518
> tpv:TP03_0179 hypothetical protein; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=444
Score = 90.9 bits (224), Expect = 9e-19, Method: Composition-based stats.
Identities = 41/70 (58%), Positives = 50/70 (71%), Gaps = 0/70 (0%)
Query 1 LGVGYEYEADVLGEEQWGWLEAQLKHSRAQAHIVVSSIQVLTGLPLVESWGLFPVSRLRL 60
G+G +++AD LGEEQW WLE+QL S A +HI+VSS QV T P+ ESWG FP S+ RL
Sbjct 230 FGIGCKHDADALGEEQWKWLESQLVDSEATSHIIVSSFQVFTYRPMGESWGHFPNSKKRL 289
Query 61 LSLFARTKPK 70
L L TKPK
Sbjct 290 LDLLEATKPK 299
> bbo:BBOV_I003300 19.m02166; hypothetical protein; K01113 alkaline
phosphatase D [EC:3.1.3.1]
Length=754
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 37/70 (52%), Positives = 46/70 (65%), Gaps = 0/70 (0%)
Query 1 LGVGYEYEADVLGEEQWGWLEAQLKHSRAQAHIVVSSIQVLTGLPLVESWGLFPVSRLRL 60
G G + D LG EQW WL+ QL S A+AHI+VSS Q+ T PL ESWGL P+++ RL
Sbjct 519 FGFGCNHPGDTLGAEQWKWLQGQLHGSTAEAHIIVSSFQIFTRFPLTESWGLLPLAKDRL 578
Query 61 LSLFARTKPK 70
+ L TKPK
Sbjct 579 VDLLLTTKPK 588
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 39 QVLTGLPLVESWGLFPVSRLRLLSLFARTKPK 70
+V T P+ ESWGL P ++ RL+ L KPK
Sbjct 180 KVFTYYPITESWGLLPQAKDRLVDLLLAAKPK 211
> pfa:PFI0605c conserved Plasmodium protein, unknown function;
K01113 alkaline phosphatase D [EC:3.1.3.1]
Length=446
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 33/65 (50%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 10 DVLGEEQWGWLEAQLKHSRAQAHIVVSSIQVLTGLPLVESWGLFPVSRLRLLSLFARTKP 69
D+LG EQW WLE +L +S A+AHI++SS Q+ + + E+WGL P S RL L +TKP
Sbjct 213 DILGNEQWKWLERELTNSNARAHIIISSTQIFSNHIINENWGLMPYSLRRLRELIKKTKP 272
Query 70 KGLML 74
KGL+
Sbjct 273 KGLLF 277
> ath:AT5G42370 hypothetical protein; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=447
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 31/74 (41%), Positives = 44/74 (59%), Gaps = 7/74 (9%)
Query 8 EADVLGEEQWGWLEAQLKHSRAQAHIVVSSIQVLTGL-----PL--VESWGLFPVSRLRL 60
+ +LG+ QW WLE +L R++ I+ SS+QV++ L PL +ESWG FP R RL
Sbjct 192 DGSILGDTQWDWLENELSGPRSEITIIGSSVQVISNLSATTGPLFYMESWGRFPKERKRL 251
Query 61 LSLFARTKPKGLML 74
L A TK G++
Sbjct 252 FKLIADTKRDGVIF 265
> tgo:TGME49_052380 hypothetical protein ; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=1222
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 38/59 (64%), Gaps = 6/59 (10%)
Query 10 DVLGEEQWGWLEAQLKHSRAQAHIVV---SSIQVLTGLPLV--ESWGLFPVSRLRLLSL 63
D+LG +QW WL+ +L+ S+ + V SSIQVL P V E+W +FP +R RLL+L
Sbjct 302 DILGNQQWRWLKEELRKSKEEGDAVTFIASSIQVLPS-PFVATEAWSIFPDARRRLLNL 359
> cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant
acid phosphatase type 5 [EC:3.1.3.2]
Length=419
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 12/22 (54%), Positives = 14/22 (63%), Gaps = 0/22 (0%)
Query 13 GEEQWGWLEAQLKHSRAQAHIV 34
EEQW WLE L+ S AQ I+
Sbjct 242 AEEQWAWLENNLEASSAQYLII 263
Lambda K H
0.321 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007182052
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40