bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
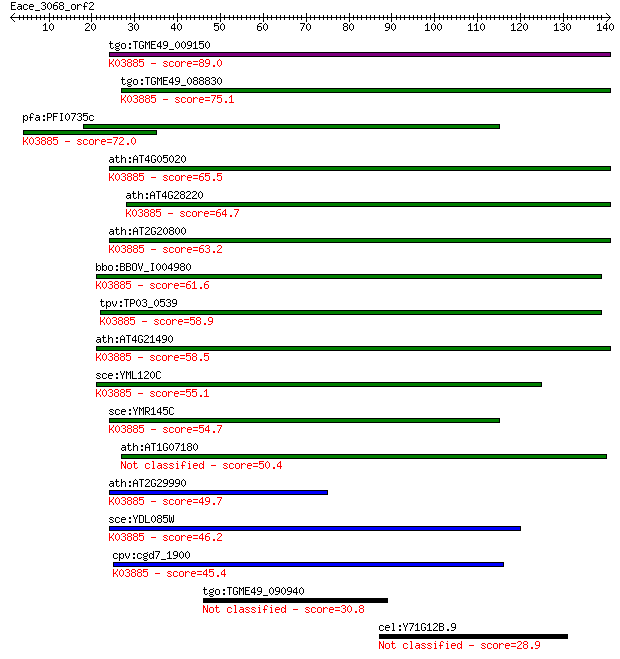
Query= Eace_3068_orf2
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_009150 mitochondrial alternative NADH dehydrogenase... 89.0 4e-18
tgo:TGME49_088830 pyridine nucleotide-disulphide oxidoreductas... 75.1 7e-14
pfa:PFI0735c NADH dehydrogenase, putative (EC:1.6.99.3); K0388... 72.0 5e-13
ath:AT4G05020 NDB2; NDB2 (NAD(P)H dehydrogenase B2); FAD bindi... 65.5 5e-11
ath:AT4G28220 NDB1; NDB1 (NAD(P)H dehydrogenase B1); NADH dehy... 64.7 1e-10
ath:AT2G20800 NDB4; NDB4 (NAD(P)H dehydrogenase B4); NADH dehy... 63.2 3e-10
bbo:BBOV_I004980 19.m02353; NADH dehydrogenase (EC:1.6.5.3); K... 61.6 8e-10
tpv:TP03_0539 NADH dehydrogenase (EC:1.6.5.3); K03885 NADH deh... 58.9 5e-09
ath:AT4G21490 NDB3; NDB3; NADH dehydrogenase; K03885 NADH dehy... 58.5 6e-09
sce:YML120C NDI1; NADH:ubiquinone oxidoreductase, transfers el... 55.1 6e-08
sce:YMR145C NDE1, NDH1; Mitochondrial external NADH dehydrogen... 54.7 8e-08
ath:AT1G07180 NDA1; NDA1 (ALTERNATIVE NAD(P)H DEHYDROGENASE 1)... 50.4 2e-06
ath:AT2G29990 NDA2; NDA2 (ALTERNATIVE NAD(P)H DEHYDROGENASE 2)... 49.7 3e-06
sce:YDL085W NDE2, NDH2; Mitochondrial external NADH dehydrogen... 46.2 3e-05
cpv:cgd7_1900 mitochondrial NADH dehydrogenase ; K03885 NADH d... 45.4 5e-05
tgo:TGME49_090940 endomembrane domain70-containing protein 30.8 1.3
cel:Y71G12B.9 lin-65; abnormal cell LINeage family member (lin... 28.9 5.2
> tgo:TGME49_009150 mitochondrial alternative NADH dehydrogenase
1 (EC:1.6.5.3); K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=618
Score = 89.0 bits (219), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 48/117 (41%), Positives = 70/117 (59%), Gaps = 7/117 (5%)
Query 24 KRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSL 83
+R +VVVVG GW++ SF++ LD ++E V ISP ++ ++TPLLP VC G LPA+AC T +
Sbjct 97 RRQKVVVVGSGWAAVSFLADLDMTRYEPVVISPRDYFTFTPLLPSVCVGTLPASACMTGV 156
Query 84 RGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYDY 140
R LL GG G +Y+ V + +V C+ A+ + WEE+YDY
Sbjct 157 RELLVRGGVP--CGSFYEGRVAEICPTEKKVRCQSTHGKAQDAR-----EWEESYDY 206
> tgo:TGME49_088830 pyridine nucleotide-disulphide oxidoreductase,
putative (EC:1.6.5.3); K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=657
Score = 75.1 bits (183), Expect = 7e-14, Method: Composition-based stats.
Identities = 40/114 (35%), Positives = 58/114 (50%), Gaps = 12/114 (10%)
Query 27 RVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSLRGL 86
+VVV+G GW+S +F LDP ++V ISP + ++TPLLP VC G L +C +R L
Sbjct 153 KVVVLGTGWASVNFFRHLDPNIYDVTVISPRNYFTFTPLLPSVCAGTLSPLSCIEPVRSL 212
Query 87 LSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYDY 140
G+ +Y+A +VD N V C S + G ++ YDY
Sbjct 213 TYRNGRKV--ADFYEAHCTDVDFKNRIVAC----------DSRQGGHFKVKYDY 254
> pfa:PFI0735c NADH dehydrogenase, putative (EC:1.6.99.3); K03885
NADH dehydrogenase [EC:1.6.99.3]
Length=533
Score = 72.0 bits (175), Expect = 5e-13, Method: Composition-based stats.
Identities = 36/112 (32%), Positives = 62/112 (55%), Gaps = 18/112 (16%)
Query 18 SNLQNKKRL----RVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGA 73
+NL+N K + +++++G GW +F+ ++D ++++V ISP + ++TPLLPC+C G
Sbjct 29 NNLKNNKDIERKEKIIILGSGWGGFNFLLNIDFKKYDVTLISPRNYFTFTPLLPCLCSGT 88
Query 74 LPAAACTTSLRGLLSFGGKAEGR-GRYYQ----------AVVDNVDLNNNRV 114
L CT S+R L K G G Y Q ++ +D+ NN+V
Sbjct 89 LSVNVCTESIRNFLR---KKNGYCGNYLQLECTDVFYEDKYINCIDIENNKV 137
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 4 RWVQCLSLCTIHKCSNLQNKKRLRVVVVGGG 34
+++ L CT+ SN + KK L V VVGGG
Sbjct 179 KFLDILEKCTLPNISNEEKKKMLHVAVVGGG 209
> ath:AT4G05020 NDB2; NDB2 (NAD(P)H dehydrogenase B2); FAD binding
/ disulfide oxidoreductase/ oxidoreductase; K03885 NADH
dehydrogenase [EC:1.6.99.3]
Length=582
Score = 65.5 bits (158), Expect = 5e-11, Method: Composition-based stats.
Identities = 38/117 (32%), Positives = 64/117 (54%), Gaps = 8/117 (6%)
Query 24 KRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSL 83
K+ +VV++G GW+ +SF+ +L+ Q+EV ISP + ++TPLLP V G + A + +
Sbjct 57 KKKKVVLLGTGWAGTSFLKNLNNSQYEVQIISPRNYFAFTPLLPSVTCGTVEARSVVEPI 116
Query 84 RGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYDY 140
R + G+ Y +A +D + +V CR L SS+G++ + YDY
Sbjct 117 RNI----GRKNVDTSYLEAECFKIDPASKKVYCRSKQGL---SSNGKK-EFSVDYDY 165
> ath:AT4G28220 NDB1; NDB1 (NAD(P)H dehydrogenase B1); NADH dehydrogenase/
disulfide oxidoreductase; K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=571
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 37/113 (32%), Positives = 57/113 (50%), Gaps = 7/113 (6%)
Query 28 VVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSLRGLL 87
VVV+G GW+ SF+ LD ++V +SP+ + ++TPLLP V G + A + S+R +
Sbjct 52 VVVLGTGWAGISFLKDLDITSYDVQVVSPQNYFAFTPLLPSVTCGTVEARSIVESVRNIT 111
Query 88 SFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYDY 140
K G ++A +D N +V CRPV +S + YDY
Sbjct 112 K---KKNGEIELWEADCFKIDHVNQKVHCRPVFKDDPEASQ----EFSLGYDY 157
> ath:AT2G20800 NDB4; NDB4 (NAD(P)H dehydrogenase B4); NADH dehydrogenase;
K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=582
Score = 63.2 bits (152), Expect = 3e-10, Method: Composition-based stats.
Identities = 37/117 (31%), Positives = 58/117 (49%), Gaps = 11/117 (9%)
Query 24 KRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSL 83
++ +VVV+G GWS SF+S L+ ++V +SP +TPLLP V G + A + +
Sbjct 62 RKKKVVVLGSGWSGYSFLSYLNNPNYDVQVVSPRNFFLFTPLLPSVTNGTVEARSIVEPI 121
Query 84 RGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYDY 140
RGL+ G Y +A +D +N ++ CR S +G+ E DY
Sbjct 122 RGLMRKKGF-----EYKEAECVKIDASNKKIHCR------SKEGSSLKGTTEFDMDY 167
> bbo:BBOV_I004980 19.m02353; NADH dehydrogenase (EC:1.6.5.3);
K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=560
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 40/134 (29%), Positives = 64/134 (47%), Gaps = 18/134 (13%)
Query 21 QNKKRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACT 80
Q + R+VV+G GWSS FV +LD +F++ +SP + ++TPLLP + G + CT
Sbjct 51 QRDHKQRIVVLGTGWSSLFFVKNLDLSKFDLQVVSPRNYFTFTPLLPKLVSGRISTKTCT 110
Query 81 TSLRGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVC---------------RPVLSL-AE 124
+ +G + A NVD ++ V C R V+++ AE
Sbjct 111 VPFSSFVQ--KHRKGSFNFVHASCVNVDPHSKLVYCVSASDPNTRVNLPYDRLVIAVGAE 168
Query 125 TSSSGERGSWEEAY 138
+++ G G E AY
Sbjct 169 SNTFGIPGVAEHAY 182
> tpv:TP03_0539 NADH dehydrogenase (EC:1.6.5.3); K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=543
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 37/133 (27%), Positives = 69/133 (51%), Gaps = 19/133 (14%)
Query 22 NKKRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTT 81
N + +V+ +G GWSS F+ +L+P+ F++ ISP + ++TPLLP + G + + T
Sbjct 38 NNTKPKVLFLGSGWSSVFFIKNLNPKLFDLTVISPRNYFTFTPLLPKILSGMVES---NT 94
Query 82 SLRGLLSFGGK-AEGRGRYYQAVVDNVDLNNNRVVCRP--------------VLSL-AET 125
S ++ + + ++ A +VD ++N V C P V+++ A+T
Sbjct 95 SAEPIIEYMRRYFRTNSQFIHAKCVDVDSDSNCVTCAPLDSGPAFSVSYDFLVIAVGAQT 154
Query 126 SSSGERGSWEEAY 138
++ G +G E AY
Sbjct 155 NTFGTKGVEEYAY 167
> ath:AT4G21490 NDB3; NDB3; NADH dehydrogenase; K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=580
Score = 58.5 bits (140), Expect = 6e-09, Method: Composition-based stats.
Identities = 36/120 (30%), Positives = 64/120 (53%), Gaps = 8/120 (6%)
Query 21 QNKKRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACT 80
+ +KR +VV++G GW+ +SF+ +L+ +EV ISP + ++TPLLP V G + A +
Sbjct 52 KTRKR-KVVLLGTGWAGASFLKTLNNSSYEVQVISPRNYFAFTPLLPSVTCGTVEARSVV 110
Query 81 TSLRGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYDY 140
+R + K + +A +D + +V CR S +S G++ ++ YDY
Sbjct 111 EPIRNI---ARKQNVEMSFLEAECFKIDPGSKKVYCR---SKQGVNSKGKK-EFDVDYDY 163
> sce:YML120C NDI1; NADH:ubiquinone oxidoreductase, transfers
electrons from NADH to ubiquinone in the respiratory chain but
does not pump protons, in contrast to the higher eukaryotic
multisubunit respiratory complex I; phosphorylated; homolog
of human AMID (EC:1.6.5.3); K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=513
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 26/104 (25%), Positives = 57/104 (54%), Gaps = 3/104 (2%)
Query 21 QNKKRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACT 80
Q+ + V+++G GW + SF+ +D +++ V ISP + +TPLLP G + +
Sbjct 49 QHSDKPNVLILGSGWGAISFLKHIDTKKYNVSIISPRSYFLFTPLLPSAPVGTVDEKSI- 107
Query 81 TSLRGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAE 124
+ +++F K +G YY+A +++ + N V + + ++++
Sbjct 108 --IEPIVNFALKKKGNVTYYEAEATSINPDRNTVTIKSLSAVSQ 149
> sce:YMR145C NDE1, NDH1; Mitochondrial external NADH dehydrogenase,
a type II NAD(P)H:quinone oxidoreductase that catalyzes
the oxidation of cytosolic NADH; Nde1p and Nde2p provide
cytosolic NADH to the mitochondrial respiratory chain (EC:1.6.5.3);
K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=560
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 46/91 (50%), Gaps = 3/91 (3%)
Query 24 KRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSL 83
KR +V++G GW S S + +LD + VV +SP + +TPLLP G + + +
Sbjct 111 KRKTLVILGSGWGSVSLLKNLDTTLYNVVVVSPRNYFLFTPLLPSTPVGTIELKSIVEPV 170
Query 84 RGLLSFGGKAEGRGRYYQAVVDNVDLNNNRV 114
R + ++ G YY+A +VD N +
Sbjct 171 R---TIARRSHGEVHYYEAEAYDVDPENKTI 198
> ath:AT1G07180 NDA1; NDA1 (ALTERNATIVE NAD(P)H DEHYDROGENASE
1); NADH dehydrogenase
Length=510
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 55/113 (48%), Gaps = 4/113 (3%)
Query 27 RVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSLRGL 86
RV+V+G GW+ + +D ++VV +SP H+ +TPLL C G L + + +
Sbjct 75 RVLVLGSGWAGCRVLKGIDTSIYDVVCVSPRNHMVFTPLLASTCVGTLEFRSVAEPISRI 134
Query 87 LSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPVLSLAETSSSGERGSWEEAYD 139
+ G Y+ A +D +N+ V C V E SS+ + ++ AYD
Sbjct 135 QPAISREPG-SYYFLANCSKLDADNHEVHCETV---TEGSSTLKPWKFKIAYD 183
> ath:AT2G29990 NDA2; NDA2 (ALTERNATIVE NAD(P)H DEHYDROGENASE
2); FAD binding / NADH dehydrogenase/ oxidoreductase; K03885
NADH dehydrogenase [EC:1.6.99.3]
Length=508
Score = 49.7 bits (117), Expect = 3e-06, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 24 KRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGAL 74
++ RVVV+G GW+ + +D ++VV +SP H+ +TPLL C G L
Sbjct 70 EKPRVVVLGSGWAGCRLMKGIDTNLYDVVCVSPRNHMVFTPLLASTCVGTL 120
> sce:YDL085W NDE2, NDH2; Mitochondrial external NADH dehydrogenase,
catalyzes the oxidation of cytosolic NADH; Nde1p and
Nde2p are involved in providing the cytosolic NADH to the mitochondrial
respiratory chain (EC:1.6.5.3); K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=545
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 44/96 (45%), Gaps = 3/96 (3%)
Query 24 KRLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSL 83
K+ +V++G GW + S + LD + V +SP +TPLLP G + + +
Sbjct 96 KKKELVILGTGWGAISLLKKLDTSLYNVTVVSPRSFFLFTPLLPSTPVGTIEMKSIVEPV 155
Query 84 RGLLSFGGKAEGRGRYYQAVVDNVDLNNNRVVCRPV 119
R S + G Y +A +VD +V+ + V
Sbjct 156 R---SIARRTPGEVHYIEAEALDVDPKAKKVMVQSV 188
> cpv:cgd7_1900 mitochondrial NADH dehydrogenase ; K03885 NADH
dehydrogenase [EC:1.6.99.3]
Length=568
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 41/91 (45%), Gaps = 2/91 (2%)
Query 25 RLRVVVVGGGWSSSSFVSSLDPRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSLR 84
R +V+++G GW LD ++ ISP ++ +TPLL + LP C +
Sbjct 62 RPKVLILGTGWGFMKLAKGLDVNSNDIKVISPNKYFCFTPLLTQIVSNRLPREVCEIPIN 121
Query 85 GLLSFGGKAEGRGRYYQAVVDNVDLNNNRVV 115
L G K +Y Q + ++D N V+
Sbjct 122 ELTYRGNKEV--IKYIQGLALDIDKENKEVI 150
> tgo:TGME49_090940 endomembrane domain70-containing protein
Length=623
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 6/43 (13%)
Query 46 PRQFEVVWISPEEHLSYTPLLPCVCGGALPAAACTTSLRGLLS 88
PRQ P++ P+L CV GGALP A T L L S
Sbjct 468 PRQI------PQQPWFMQPVLSCVVGGALPFGAMFTELFFLFS 504
> cel:Y71G12B.9 lin-65; abnormal cell LINeage family member (lin-65)
Length=728
Score = 28.9 bits (63), Expect = 5.2, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 28/45 (62%), Gaps = 8/45 (17%)
Query 87 LSFGGKAEGRGRYYQAVVDNVDLNNNRV-VCRPVLSLAETSSSGE 130
L+ GG+A Q ++DN +++N + +C P++S A T SSG+
Sbjct 552 LNNGGRA-------QPLIDNTRVHDNTIMLCVPLVSTANTISSGD 589
Lambda K H
0.320 0.134 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40