bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3072_orf2
Length=160
Score E
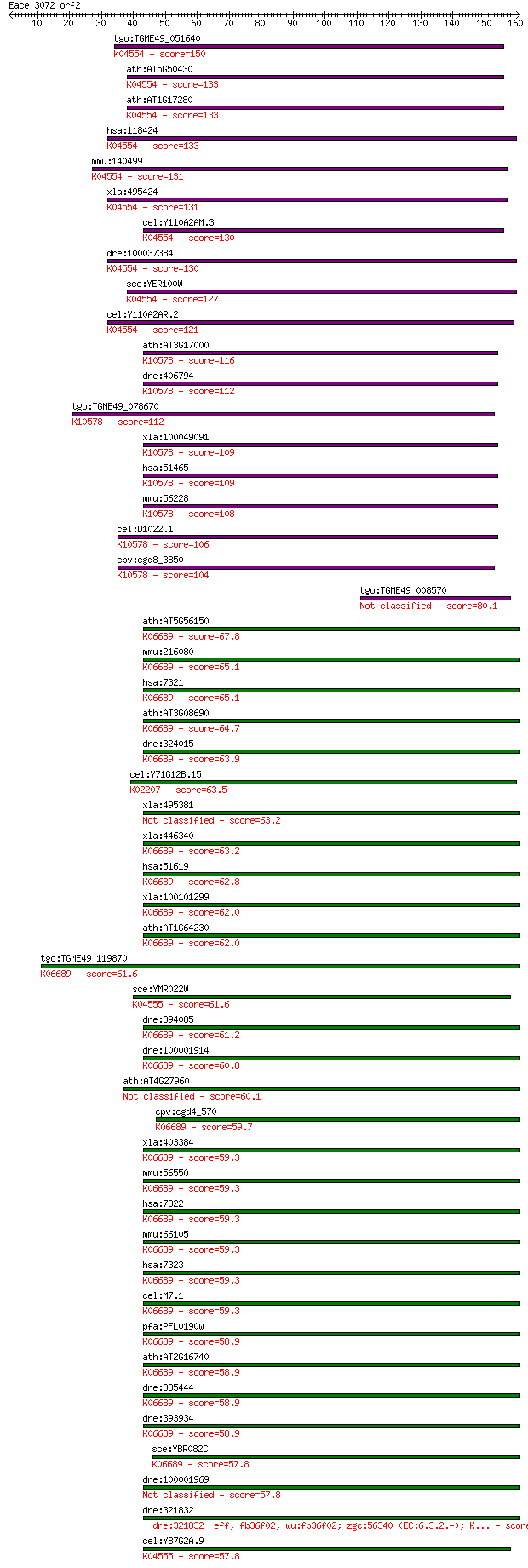
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_051640 ubiquitin-conjugating enzyme E2, putative (E... 150 1e-36
ath:AT5G50430 UBC33; UBC33 (ubiquitin-conjugating enzyme 33); ... 133 2e-31
ath:AT1G17280 UBC34; UBC34 (ubiquitin-conjugating enzyme 34); ... 133 3e-31
hsa:118424 UBE2J2, NCUBE-2, NCUBE2, PRO2121; ubiquitin-conjuga... 133 3e-31
mmu:140499 Ube2j2, 1200007B18Rik, 2400008G19Rik, 5730472G04Rik... 131 8e-31
xla:495424 ube2j2; ubiquitin-conjugating enzyme E2, J2 (UBC6 h... 131 1e-30
cel:Y110A2AM.3 ubc-26; UBiquitin Conjugating enzyme family mem... 130 1e-30
dre:100037384 ube2j2, zgc:162164; ubiquitin-conjugating enzyme... 130 2e-30
sce:YER100W UBC6, DOA2; Ubc6p (EC:6.3.2.19); K04554 ubiquitin-... 127 2e-29
cel:Y110A2AR.2 ubc-15; UBiquitin Conjugating enzyme family mem... 121 1e-27
ath:AT3G17000 UBC32; UBC32 (ubiquitin-conjugating enzyme 32); ... 116 4e-26
dre:406794 zgc:63554 (EC:6.3.2.19); K10578 ubiquitin-conjugati... 112 4e-25
tgo:TGME49_078670 ubiquitin-conjugating enzyme E2, putative (E... 112 4e-25
xla:100049091 ube2j1; ubiquitin-conjugating enzyme E2, J1 (UBC... 109 4e-24
hsa:51465 UBE2J1, HSPC153, HSPC205, HSU93243, MGC12555, NCUBE-... 109 5e-24
mmu:56228 Ube2j1, 0710008M05Rik, 1110030I22Rik, NCUBE-1, Ncube... 108 6e-24
cel:D1022.1 ubc-6; UBiquitin Conjugating enzyme family member ... 106 3e-23
cpv:cgd8_3850 Ubc6p like ubiquiting conjugating enzyme E2, pos... 104 1e-22
tgo:TGME49_008570 hypothetical protein 80.1 3e-15
ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30); ... 67.8 1e-11
mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzy... 65.1 1e-10
hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiqui... 65.1 1e-10
ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11); ... 64.7 1e-10
dre:324015 ube2d1, wu:fc16h06, zgc:73096; ubiquitin-conjugatin... 63.9 2e-10
cel:Y71G12B.15 ubc-3; UBiquitin Conjugating enzyme family memb... 63.5 3e-10
xla:495381 ube2d1, e2(17)kb1, sft, ubc4/5, ubch5, ubch5a; ubiq... 63.2 4e-10
xla:446340 ube2d2, MGC81261; ubiquitin-conjugating enzyme E2D ... 63.2 4e-10
hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzy... 62.8 4e-10
xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating ... 62.0 8e-10
ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative; K... 62.0 9e-10
tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containi... 61.6 1e-09
sce:YMR022W UBC7, QRI8; Ubc7p (EC:6.3.2.19); K04555 ubiquitin-... 61.6 1e-09
dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689 ub... 61.2 1e-09
dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjuga... 60.8 2e-09
ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubi... 60.1 3e-09
cpv:cgd4_570 ubiquitin-conjugating enzyme ; K06689 ubiquitin-c... 59.7 4e-09
xla:403384 ube2d3, ube2d2, ube2d3.1, xubc4; ubiquitin-conjugat... 59.3 5e-09
mmu:56550 Ube2d2, 1500034D03Rik, Ubc2e, ubc4; ubiquitin-conjug... 59.3 5e-09
hsa:7322 UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B; ubiqu... 59.3 5e-09
mmu:66105 Ube2d3, 1100001F19Rik, 9430029A22Rik, AA414951; ubiq... 59.3 5e-09
hsa:7323 UBE2D3, E2(17)KB3, MGC43926, MGC5416, UBC4/5, UBCH5C;... 59.3 5e-09
cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquit... 59.3 6e-09
pfa:PFL0190w ubiquitin conjugating enzyme E2, putative (EC:6.3... 58.9 6e-09
ath:AT2G16740 UBC29; UBC29 (ubiquitin-conjugating enzyme 29); ... 58.9 6e-09
dre:335444 ube2d2, wu:fj13d01, zgc:73200; ubiquitin-conjugatin... 58.9 7e-09
dre:393934 MGC55886, Ube2d2, zgc:77149; zgc:55886 (EC:6.3.2.19... 58.9 7e-09
sce:YBR082C UBC4; Ubc4p (EC:6.3.2.19); K06689 ubiquitin-conjug... 57.8 2e-08
dre:100001969 ubiquitin-conjugating enzyme E2D 2-like 57.8 2e-08
dre:321832 eff, fb36f02, wu:fb36f02; zgc:56340 (EC:6.3.2.-); K... 57.8 2e-08
cel:Y87G2A.9 ubc-14; UBiquitin Conjugating enzyme family membe... 57.8 2e-08
> tgo:TGME49_051640 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=275
Score = 150 bits (380), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 68/126 (53%), Positives = 91/126 (72%), Gaps = 4/126 (3%)
Query 34 SSSKMSSKGAV---DRLRREWKSLQRES-LPNVEARPQEDDFLTWIFLIHDLPEDSSFAG 89
S K +GA RL RE+ LQR+ +P+ + +P +D LTW F++HDLP DS + G
Sbjct 2 SRDKGGGRGAFLGPGRLAREFSLLQRQGGVPHAQLQPDMNDTLTWHFVLHDLPADSPYHG 61
Query 90 GFYMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGF 149
G Y GK+VFP YPF PPSI+MLTP+GRFE N+ +C+S + +HPE WNPSW++ETL+T F
Sbjct 62 GVYHGKLVFPPNYPFEPPSIFMLTPSGRFEVNKRICMSMSDFHPESWNPSWRLETLVTAF 121
Query 150 VSFMLD 155
+SFMLD
Sbjct 122 LSFMLD 127
> ath:AT5G50430 UBC33; UBC33 (ubiquitin-conjugating enzyme 33);
ubiquitin-protein ligase; K04554 ubiquitin-conjugating enzyme
E2 J2 [EC:6.3.2.19]
Length=243
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 55/118 (46%), Positives = 81/118 (68%), Gaps = 1/118 (0%)
Query 38 MSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVV 97
M+ K + RL++E+++L +E + +V ARP +D L W +++ E + FAGGFY GK+
Sbjct 1 MAEKACIKRLQKEYRALCKEPVSHVVARPSPNDILEWHYVLEG-SEGTPFAGGFYYGKIK 59
Query 98 FPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLD 155
FP YP+ PP I M TPNGRF T + +C+S + +HPE WNP W V ++LTG +SFM+D
Sbjct 60 FPPEYPYKPPGITMTTPNGRFVTQKKICLSMSDFHPESWNPMWSVSSILTGLLSFMMD 117
> ath:AT1G17280 UBC34; UBC34 (ubiquitin-conjugating enzyme 34);
ubiquitin-protein ligase; K04554 ubiquitin-conjugating enzyme
E2 J2 [EC:6.3.2.19]
Length=237
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 55/118 (46%), Positives = 81/118 (68%), Gaps = 1/118 (0%)
Query 38 MSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVV 97
M+ K + RL++E+++L +E + +V ARP +D L W +++ E + FAGGFY GK+
Sbjct 1 MAEKACIKRLQKEYRALCKEPVSHVVARPSPNDILEWHYVLEG-SEGTPFAGGFYYGKIK 59
Query 98 FPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLD 155
FP YP+ PP I M TPNGRF T + +C+S + +HPE WNP W V ++LTG +SFM+D
Sbjct 60 FPPEYPYKPPGITMTTPNGRFMTQKKICLSMSDFHPESWNPMWSVSSILTGLLSFMMD 117
> hsa:118424 UBE2J2, NCUBE-2, NCUBE2, PRO2121; ubiquitin-conjugating
enzyme E2, J2 (UBC6 homolog, yeast) (EC:6.3.2.19); K04554
ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=259
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 55/128 (42%), Positives = 89/128 (69%), Gaps = 1/128 (0%)
Query 32 SSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGF 91
SS+SSK + A RL++++ ++++ +P + A P + L W +++ PE + + GG+
Sbjct 2 SSTSSKRAPTTATQRLKQDYLRIKKDPVPYICAEPLPSNILEWHYVVRG-PEMTPYEGGY 60
Query 92 YMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVS 151
Y GK++FP +PF PPSIYM+TPNGRF+ N LC+S T +HP+ WNP+W V T+LTG +S
Sbjct 61 YHGKLIFPREFPFKPPSIYMITPNGRFKCNTRLCLSITDFHPDTWNPAWSVSTILTGLLS 120
Query 152 FMLDENAS 159
FM+++ +
Sbjct 121 FMVEKGPT 128
> mmu:140499 Ube2j2, 1200007B18Rik, 2400008G19Rik, 5730472G04Rik,
AL022923, NCUBE-2, Ubc6, Ubc6p; ubiquitin-conjugating enzyme
E2, J2 homolog (yeast) (EC:6.3.2.19); K04554 ubiquitin-conjugating
enzyme E2 J2 [EC:6.3.2.19]
Length=271
Score = 131 bits (330), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 54/130 (41%), Positives = 90/130 (69%), Gaps = 1/130 (0%)
Query 27 SSSSSSSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSS 86
S+ S++S+K + A RL++++ ++++ +P + A P + L W +++ PE +
Sbjct 9 SARKMSNNSNKRAPTTATQRLKQDYLRIKKDPVPYICAEPLPSNILEWHYVVRG-PEMTP 67
Query 87 FAGGFYMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLL 146
+ GG+Y GK++FP +PF PPSIYM+TPNGRF+ N LC+S T +HP+ WNP+W V T+L
Sbjct 68 YEGGYYHGKLIFPREFPFKPPSIYMITPNGRFKCNTRLCLSITDFHPDTWNPAWSVSTIL 127
Query 147 TGFVSFMLDE 156
TG +SFM+++
Sbjct 128 TGLLSFMVEK 137
> xla:495424 ube2j2; ubiquitin-conjugating enzyme E2, J2 (UBC6
homolog); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=259
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 54/125 (43%), Positives = 88/125 (70%), Gaps = 1/125 (0%)
Query 32 SSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGF 91
S++S+K + A RL++++ ++++ +P + A P + L W +++ PE + + GG+
Sbjct 2 SNNSNKRAPTTATQRLKQDYLRIKKDPVPYICAEPLPSNILEWHYVVRG-PEMTPYEGGY 60
Query 92 YMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVS 151
Y GK+VFP +PF PPSIYM+TPNGRF+ N LC+S T +HP+ WNP+W V T+LTG +S
Sbjct 61 YHGKLVFPREFPFKPPSIYMITPNGRFKCNTRLCLSITDFHPDTWNPAWSVSTILTGLLS 120
Query 152 FMLDE 156
FM+++
Sbjct 121 FMVEK 125
> cel:Y110A2AM.3 ubc-26; UBiquitin Conjugating enzyme family member
(ubc-26); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=203
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 54/113 (47%), Positives = 82/113 (72%), Gaps = 1/113 (0%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ RL+++++ L +E +P ++A P E + L W ++I P+ + + GG YMGK++FP +
Sbjct 9 ALQRLKKDYQRLLKEPVPFMKAAPLETNILEWRYIIIGAPK-TPYEGGIYMGKLLFPKDF 67
Query 103 PFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLD 155
PF PP+I MLTPNGRF+TN LC+S + YHP+ WNP+W V T++TG +SFM D
Sbjct 68 PFKPPAILMLTPNGRFQTNTRLCLSISDYHPDTWNPAWTVSTIITGLMSFMND 120
> dre:100037384 ube2j2, zgc:162164; ubiquitin-conjugating enzyme
E2, J2 (UBC6 homolog, yeast) (EC:6.3.2.19); K04554 ubiquitin-conjugating
enzyme E2 J2 [EC:6.3.2.19]
Length=259
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 88/128 (68%), Gaps = 1/128 (0%)
Query 32 SSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGF 91
S++ +K + A RL++++ ++++ +P + A P + L W +L+ PE + + GG+
Sbjct 2 SNNLNKRAPTTATQRLKQDYLRIKKDPVPYICAEPLPSNILEWHYLVRG-PEKTPYEGGY 60
Query 92 YMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVS 151
Y GK++FP +PF PPSIYM+TPNGRF+ N LC+S T +HP+ WNP+W V T+LTG +S
Sbjct 61 YHGKLIFPREFPFKPPSIYMITPNGRFKCNTRLCLSITDFHPDTWNPAWSVSTILTGLLS 120
Query 152 FMLDENAS 159
FM+++ +
Sbjct 121 FMVEKGPT 128
> sce:YER100W UBC6, DOA2; Ubc6p (EC:6.3.2.19); K04554 ubiquitin-conjugating
enzyme E2 J2 [EC:6.3.2.19]
Length=250
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 55/122 (45%), Positives = 79/122 (64%), Gaps = 1/122 (0%)
Query 38 MSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVV 97
M++K A RL +E+K + P + ARP ED+ L W ++I P D+ + GG Y G +
Sbjct 1 MATKQAHKRLTKEYKLMVENPPPYILARPNEDNILEWHYIITG-PADTPYKGGQYHGTLT 59
Query 98 FPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDEN 157
FP+ YP+ PP+I M+TPNGRF+ N LC+S + YHP+ WNP W V T+L G +SFM +
Sbjct 60 FPSDYPYKPPAIRMITPNGRFKPNTRLCLSMSDYHPDTWNPGWSVSTILNGLLSFMTSDE 119
Query 158 AS 159
A+
Sbjct 120 AT 121
> cel:Y110A2AR.2 ubc-15; UBiquitin Conjugating enzyme family member
(ubc-15); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=218
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/127 (44%), Positives = 84/127 (66%), Gaps = 2/127 (1%)
Query 32 SSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGF 91
S ++ +S AV RL++++ L ++ + ++A P ED+ L W + + P D+ F GG+
Sbjct 9 SVPAAGTASSSAVRRLQKDYAKLMQDPVDGIKALPNEDNILEWHYCLRGSP-DTPFYGGY 67
Query 92 YMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVS 151
Y GKV+F +P+SPP+I M+TPNGRF+TN LC+S + YHPE WNP W V +L G S
Sbjct 68 YWGKVIFKENFPWSPPAITMITPNGRFQTNTRLCLSISDYHPESWNPGWTVSAILIGLHS 127
Query 152 FMLDENA 158
FM +EN+
Sbjct 128 FM-NENS 133
> ath:AT3G17000 UBC32; UBC32 (ubiquitin-conjugating enzyme 32);
ubiquitin-protein ligase; K10578 ubiquitin-conjugating enzyme
E2 J1 [EC:6.3.2.19]
Length=309
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 51/111 (45%), Positives = 68/111 (61%), Gaps = 1/111 (0%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
AV R+ +E K +Q + + P E++ W F I P D+ F GG Y G++ PA Y
Sbjct 12 AVKRILQEVKEMQANPSDDFMSLPLEENIFEWQFAIRG-PGDTEFEGGIYHGRIQLPADY 70
Query 103 PFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFM 153
PF PPS +LTPNGRFETN +C+S ++YHPE W PSW V T L ++FM
Sbjct 71 PFKPPSFMLLTPNGRFETNTKICLSISNYHPEHWQPSWSVRTALVALIAFM 121
> dre:406794 zgc:63554 (EC:6.3.2.19); K10578 ubiquitin-conjugating
enzyme E2 J1 [EC:6.3.2.19]
Length=314
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 50/111 (45%), Positives = 66/111 (59%), Gaps = 2/111 (1%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
AV RL +E L R+ + A+P ED+ W F + P DS F GG Y G++V P Y
Sbjct 11 AVKRLMKEAAEL-RDPTEHYHAQPLEDNLFEWHFSVRG-PPDSDFDGGVYHGRIVLPPEY 68
Query 103 PFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFM 153
P PPSI +LTPNGRFE + +C+S + +HPE W PSW + T L + FM
Sbjct 69 PMKPPSIILLTPNGRFEVGKKICLSISGHHPETWQPSWSIRTALIAIIGFM 119
> tgo:TGME49_078670 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=201
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 50/133 (37%), Positives = 79/133 (59%), Gaps = 2/133 (1%)
Query 21 SSTSNSSSSSSSSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARP-QEDDFLTWIFLIH 79
SN + ++++ ++ + R+ RE++ +QR P+ A P Q ++ W F +
Sbjct 2 QGVSNPGRTGTNANLGYTATAQCIARILREFREIQRTPSPHWCANPLQIEEPYEWHFTLR 61
Query 80 DLPEDSSFAGGFYMGKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPS 139
P+DS F GG Y G++V P YPF+PP++ MLT NGRFE + +C+S + YHPELW P+
Sbjct 62 G-PQDSHFEGGLYHGRIVLPKNYPFAPPNLVMLTQNGRFEVGKKVCLSASSYHPELWQPA 120
Query 140 WKVETLLTGFVSF 152
W + TLL +F
Sbjct 121 WGIRTLLDALCAF 133
> xla:100049091 ube2j1; ubiquitin-conjugating enzyme E2, J1 (UBC6
homolog); K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=302
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 49/111 (44%), Positives = 65/111 (58%), Gaps = 2/111 (1%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
AV RL +E L R+ + A+P ED+ W F + P DS F GG Y G++V P Y
Sbjct 11 AVKRLMKEAAEL-RDPTDHYHAQPLEDNLFEWHFTVRG-PPDSDFDGGVYHGRIVLPPEY 68
Query 103 PFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFM 153
P PPSI +LT NGRFE + +C+S + +HPE W PSW + T L + FM
Sbjct 69 PMKPPSIILLTANGRFEVGKKICLSISGHHPETWQPSWSIRTALLAIIGFM 119
> hsa:51465 UBE2J1, HSPC153, HSPC205, HSU93243, MGC12555, NCUBE-1,
NCUBE1, Ubc6p; ubiquitin-conjugating enzyme E2, J1 (UBC6
homolog, yeast) (EC:6.3.2.19); K10578 ubiquitin-conjugating
enzyme E2 J1 [EC:6.3.2.19]
Length=318
Score = 109 bits (272), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 48/111 (43%), Positives = 65/111 (58%), Gaps = 2/111 (1%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
AV RL +E L ++ + A+P ED+ W F + P DS F GG Y G++V P Y
Sbjct 11 AVKRLMKEAAEL-KDPTDHYHAQPLEDNLFEWHFTVRG-PPDSDFDGGVYHGRIVLPPEY 68
Query 103 PFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFM 153
P PPSI +LT NGRFE + +C+S + +HPE W PSW + T L + FM
Sbjct 69 PMKPPSIILLTANGRFEVGKKICLSISGHHPETWQPSWSIRTALLAIIGFM 119
> mmu:56228 Ube2j1, 0710008M05Rik, 1110030I22Rik, NCUBE-1, Ncube,
Ncube1, Ubc6p; ubiquitin-conjugating enzyme E2, J1 (EC:6.3.2.19);
K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=318
Score = 108 bits (271), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 48/111 (43%), Positives = 65/111 (58%), Gaps = 2/111 (1%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
AV RL +E L ++ + A+P ED+ W F + P DS F GG Y G++V P Y
Sbjct 11 AVKRLMKEAAEL-KDPTDHYHAQPLEDNLFEWHFTVRG-PPDSDFDGGVYHGRIVLPPEY 68
Query 103 PFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFM 153
P PPSI +LT NGRFE + +C+S + +HPE W PSW + T L + FM
Sbjct 69 PMKPPSIILLTANGRFEVGKKICLSISGHHPETWQPSWSIRTALLAIIGFM 119
> cel:D1022.1 ubc-6; UBiquitin Conjugating enzyme family member
(ubc-6); K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=314
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 50/120 (41%), Positives = 71/120 (59%), Gaps = 3/120 (2%)
Query 35 SSKMSSKGA-VDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYM 93
S + ++K A V RL +E L R+ A+P ED+ W F I + F GG Y
Sbjct 2 SEQYNTKNAGVRRLMKEAMEL-RQPTEMYHAQPMEDNLFEWHFTIRGT-LGTDFEGGIYH 59
Query 94 GKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFM 153
G+++FPA YP PP++ +LTPNGRFE N+ +C+S + YHPE W PSW + T L + F+
Sbjct 60 GRIIFPADYPMKPPNLILLTPNGRFELNKKVCLSISGYHPETWLPSWSIRTALLALIGFL 119
> cpv:cgd8_3850 Ubc6p like ubiquiting conjugating enzyme E2, possible
transmembrane domain at C ; K10578 ubiquitin-conjugating
enzyme E2 J1 [EC:6.3.2.19]
Length=368
Score = 104 bits (259), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 49/119 (41%), Positives = 73/119 (61%), Gaps = 2/119 (1%)
Query 35 SSKMSSKGAVDRLRREWKSLQRESLPNVEARP-QEDDFLTWIFLIHDLPEDSSFAGGFYM 93
SS ++S + R+ RE++ +Q+E A P D+ W F I P + F GG Y
Sbjct 43 SSGITSVQCLSRILREYREIQKEPSSYWCAFPINMDEPYEWHFTIKG-PAGTEFEGGMYH 101
Query 94 GKVVFPATYPFSPPSIYMLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSF 152
G+++ P +YPFSPPS+ MLT NGRFE + +C+S ++YHPELW P+W + T+L +F
Sbjct 102 GRIILPHSYPFSPPSLMMLTGNGRFEVGKKVCLSASNYHPELWQPAWGIRTMLDALHAF 160
> tgo:TGME49_008570 hypothetical protein
Length=675
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 30/47 (63%), Positives = 40/47 (85%), Gaps = 0/47 (0%)
Query 111 MLTPNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDEN 157
M+TP+GRF NR LC+S + +HPE WNPSW+VET+LTG +SFM+DE+
Sbjct 1 MMTPSGRFAPNRRLCLSISDFHPETWNPSWRVETILTGLLSFMIDED 47
> ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/122 (33%), Positives = 57/122 (46%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A R+ +E + LQR+ + A P DD W I P DS FAGG ++ + FP Y
Sbjct 2 ASKRINKELRDLQRDPPVSCSAGPTGDDMFQWQATIMG-PADSPFAGGVFLVTIHFPPDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ E W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFRTKVYHPN--INSNGSICLDILK---EQWSPALTVSKVLLSICSLLTDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzyme
E2D 1, UBC4/5 homolog (yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 56/122 (45%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ + A P DD W I P DS++ GG + V FP Y
Sbjct 2 ALKRIQKELSDLQRDPPAHCSAGPVGDDLFHWQATIMG-PPDSAYQGGVFFLTVHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP I T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKIAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 56/122 (45%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ + A P DD W I P DS++ GG + V FP Y
Sbjct 2 ALKRIQKELSDLQRDPPAHCSAGPVGDDLFHWQATIMG-PPDSAYQGGVFFLTVHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP I T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKIAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 57/122 (46%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A R+ +E K LQ++ N A P +D W I PE S +AGG ++ + FP Y
Sbjct 2 ASKRILKELKDLQKDPPSNCSAGPVAEDMFHWQATIMGPPE-SPYAGGVFLVSIHFPPDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ E W+P+ + +L S + D N
Sbjct 61 PFKPPKVSFKTKVYHPN--INSNGSICLDILK---EQWSPALTISKVLLSICSLLTDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> dre:324015 ube2d1, wu:fc16h06, zgc:73096; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 55/122 (45%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E + LQR+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIQKELQDLQRDPPAQCSAGPVGDDLFHWQATIMG-PSDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> cel:Y71G12B.15 ubc-3; UBiquitin Conjugating enzyme family member
(ubc-3); K02207 ubiquitin-conjugating enzyme E2 R [EC:6.3.2.19]
Length=243
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 63/135 (46%), Gaps = 17/135 (12%)
Query 39 SSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVF 98
+S GA+ L E K+LQ + + ED+ W I+ P+ + + GG++ + F
Sbjct 7 TSSGALRALTMELKNLQSQPVEGFTIDVNEDNLFVWTVGIYGPPK-TLYQGGYFKASIRF 65
Query 99 PATYPFSPPSIYMLT----PNGRFETNRSLCVSFTH----------YHPELWNPSWKVET 144
P+ YP+SPPS+ T PN N LC+S H E WNP+ V T
Sbjct 66 PSNYPYSPPSMKFTTKVWHPN--VYENGDLCISILHSPIDDPQSGELACERWNPTQSVRT 123
Query 145 LLTGFVSFMLDENAS 159
+L +S + + N S
Sbjct 124 ILLSVISLLNEPNTS 138
> xla:495381 ube2d1, e2(17)kb1, sft, ubc4/5, ubch5, ubch5a; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog)
Length=147
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIQKELNDLQRDPPAQCSAGPVGDDLFHWQATIMG-PTDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> xla:446340 ube2d2, MGC81261; ubiquitin-conjugating enzyme E2D
2 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIQKELNDLQRDPPAQCSAGPVGDDLFHWQATIMG-PTDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzyme
E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIQKELTDLQRDPPAQCSAGPVGDDLFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ A P +D W I P DS F GG + + FP Y
Sbjct 2 ALKRIQKELMDLQRDPPAQCSAGPVGEDLFHWQATIMG-PNDSPFQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 57/122 (46%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A R+ +E K LQ++ + A P +D W I P DS ++GG ++ + FP Y
Sbjct 2 ASKRILKELKDLQKDPPTSCSAGPVAEDMFHWQATIMG-PSDSPYSGGVFLVTIHFPPDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ E W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFRTKVFHPN--VNSNGSICLDILK---EQWSPALTISKVLLSICSLLTDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containing
protein (EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=345
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/154 (27%), Positives = 67/154 (43%), Gaps = 22/154 (14%)
Query 11 YSASSSRIRTSSTSNSSSSSSSSSSSKMSSKGAVDRLRREWKSLQRESLPNVEARPQEDD 70
+ ASS R+R+ T S+ A+ R+ +E L ++ N A P DD
Sbjct 180 HPASSPRLRSIHTDTEVSTM------------ALKRINKELNDLSKDPPTNCSAGPVGDD 227
Query 71 FLTWIFLIHDLPEDSSFAGGFYMGKVVFPATYPFSPPSIYMLT----PNGRFETNRSLCV 126
W I PEDS ++GG + + FP+ YPF PP + T PN + ++C+
Sbjct 228 MFHWQATIMG-PEDSPYSGGVFFLNIHFPSDYPFKPPKVNFTTKIYHPN--INSQGAICL 284
Query 127 SFTHYHPELWNPSWKVETLLTGFVSFMLDENASD 160
+ W+P+ + +L S + D N D
Sbjct 285 DILK---DQWSPALTISKVLLSISSLLTDPNPDD 315
> sce:YMR022W UBC7, QRI8; Ubc7p (EC:6.3.2.19); K04555 ubiquitin-conjugating
enzyme E2 G2 [EC:6.3.2.19]
Length=165
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 63/133 (47%), Gaps = 18/133 (13%)
Query 40 SKGAVDRLRREWKSLQRESLPNVEARPQ-EDDFLTWIFLIHDLPEDSSFAGGFYMGKVVF 98
SK A RL +E + L ++S P + A P+ E++ W LI P D+ +A G + K+ F
Sbjct 2 SKTAQKRLLKELQQLIKDSPPGIVAGPKSENNIFIWDCLIQG-PPDTPYADGVFNAKLEF 60
Query 99 PATYPFSPPSIY----MLTPNGRFETNRSLCVSFTH----------YHPELWNPSWKVET 144
P YP SPP + +L PN N +C+S H E W+P VE
Sbjct 61 PKDYPLSPPKLTFTPSILHPN--IYPNGEVCISILHSPGDDPNMYELAEERWSPVQSVEK 118
Query 145 LLTGFVSFMLDEN 157
+L +S + + N
Sbjct 119 ILLSVMSMLSEPN 131
> dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 55/122 (45%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E + LQR+ A P +D W I P DS + GG + + FP Y
Sbjct 2 ALKRIQKELQDLQRDPPSQCSAGPLGEDLFHWQATIMG-PGDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E LQR+ A P +D W I P DS + GG + + FP Y
Sbjct 2 ALKRIQKELTDLQRDPPAQCSAGPVGEDLFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ V +L S + D N
Sbjct 61 PFKPPKVAFTTKIYHPN--INSNGSICLDILRSQ---WSPALTVSKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubiquitin-protein
ligase
Length=178
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 61/128 (47%), Gaps = 14/128 (10%)
Query 37 KMSSKGAVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKV 96
+M+SK R+ +E K LQ++ + A P +D W I P DS ++GG ++ +
Sbjct 30 EMASK----RILKELKDLQKDPPTSCSAGPVAEDMFHWQATIMG-PSDSPYSGGVFLVTI 84
Query 97 VFPATYPFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSF 152
FP YPF PP + T PN +N S+C+ E W+P+ + +L S
Sbjct 85 HFPPDYPFKPPKVAFRTKVFHPN--INSNGSICLDILK---EQWSPALTISKVLLSICSL 139
Query 153 MLDENASD 160
+ D N D
Sbjct 140 LTDPNPDD 147
> cpv:cgd4_570 ubiquitin-conjugating enzyme ; K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=161
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 54/116 (46%), Gaps = 6/116 (5%)
Query 47 LRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATYPFSP 106
+ +E L R+ N A P DD W I P+DSS+AGG + + FP+ YPF P
Sbjct 20 IIQELSDLNRDPPTNCSAGPIGDDMFHWQATIMG-PDDSSYAGGVFFLNIQFPSDYPFKP 78
Query 107 PSIYMLTP--NGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENASD 160
P + T + +N ++C+ E W+P+ + +L S + D N D
Sbjct 79 PKVNFTTKIYHCNINSNGAICLDILK---EQWSPALTISKVLLSISSLLTDANPDD 131
> xla:403384 ube2d3, ube2d2, ube2d3.1, xubc4; ubiquitin-conjugating
enzyme E2D 3 (UBC4/5 homolog) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIHKELNDLARDPPAQCSAGPVGDDMFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> mmu:56550 Ube2d2, 1500034D03Rik, Ubc2e, ubc4; ubiquitin-conjugating
enzyme E2D 2 (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIHKELNDLARDPPAQCSAGPVGDDMFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> hsa:7322 UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B; ubiquitin-conjugating
enzyme E2D 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIHKELNDLARDPPAQCSAGPVGDDMFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> mmu:66105 Ube2d3, 1100001F19Rik, 9430029A22Rik, AA414951; ubiquitin-conjugating
enzyme E2D 3 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRINKELSDLARDPPAQCSAGPVGDDMFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> hsa:7323 UBE2D3, E2(17)KB3, MGC43926, MGC5416, UBC4/5, UBCH5C;
ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast)
(EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme E2 D/E
[EC:6.3.2.19]
Length=147
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRINKELSDLARDPPAQCSAGPVGDDMFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 59.3 bits (142), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E + L R+ A P DD W I PE S + GG + + FP Y
Sbjct 2 ALKRIQKELQDLGRDPPAQCSAGPVGDDLFHWQATIMGPPE-SPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> pfa:PFL0190w ubiquitin conjugating enzyme E2, putative (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 54/122 (44%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E + L ++ N A P DD W I P DS + G Y + FP Y
Sbjct 2 ALKRITKELQDLNKDPPTNCSAGPIGDDLFFWQATIMG-PGDSPYENGVYFLNIKFPPDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP I T PN T ++C+ + W+P+ + +L S + D NA
Sbjct 61 PFKPPKIIFTTKIYHPN--INTAGAICLDILK---DQWSPALTISKVLLSISSLLTDPNA 115
Query 159 SD 160
D
Sbjct 116 DD 117
> ath:AT2G16740 UBC29; UBC29 (ubiquitin-conjugating enzyme 29);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A R+ +E K LQR+ + A P +D W I P +S ++GG ++ + FP Y
Sbjct 2 ATRRILKELKELQRDPPVSCSAGPTGEDMFHWQATIMG-PNESPYSGGVFLVNIHFPPDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N ++C+ + W+P+ + +L S + D N
Sbjct 61 PFKPPKVVFRTKVFHPN--INSNGNICLDILK---DQWSPALTISKVLLSICSLLTDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> dre:335444 ube2d2, wu:fj13d01, zgc:73200; ubiquitin-conjugating
enzyme E2D 2 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIHKELTDLGRDPPAQCSAGPVGDDLFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> dre:393934 MGC55886, Ube2d2, zgc:77149; zgc:55886 (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+ +E L R+ A P DD W I P DS + GG + + FP Y
Sbjct 2 ALKRIHKELHDLGRDPPAQCSAGPVGDDMFHWQATIMG-PNDSPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> sce:YBR082C UBC4; Ubc4p (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 53/119 (44%), Gaps = 10/119 (8%)
Query 46 RLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATYPFS 105
R+ +E L+R+ + A P DD W I P DS +AGG + + FP YPF
Sbjct 6 RIAKELSDLERDPPTSCSAGPVGDDLYHWQASIMG-PADSPYAGGVFFLSIHFPTDYPFK 64
Query 106 PPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENASD 160
PP I T PN N ++C+ + W+P+ + +L S + D N D
Sbjct 65 PPKISFTTKIYHPN--INANGNICLDILK---DQWSPALTLSKVLLSICSLLTDANPDD 118
> dre:100001969 ubiquitin-conjugating enzyme E2D 2-like
Length=147
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 53/122 (43%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E L R+ A P DD W I P +S + GG + + FP Y
Sbjct 2 ALKRIQKELTDLARDPPAQCSAGPVGDDVFHWQATIMG-PNESPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> dre:321832 eff, fb36f02, wu:fb36f02; zgc:56340 (EC:6.3.2.-);
K01932 [EC:6.3.2.-]
Length=147
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 53/122 (43%), Gaps = 10/122 (8%)
Query 43 AVDRLRREWKSLQRESLPNVEARPQEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPATY 102
A+ R+++E L R+ A P DD W I P +S + GG + + FP Y
Sbjct 2 ALKRIQKELTDLARDPPAQCSAGPVGDDVFHWQATIMG-PNESPYQGGVFFLTIHFPTDY 60
Query 103 PFSPPSIYMLT----PNGRFETNRSLCVSFTHYHPELWNPSWKVETLLTGFVSFMLDENA 158
PF PP + T PN +N S+C+ W+P+ + +L S + D N
Sbjct 61 PFKPPKVAFTTRIYHPN--INSNGSICLDILRSQ---WSPALTISKVLLSICSLLCDPNP 115
Query 159 SD 160
D
Sbjct 116 DD 117
> cel:Y87G2A.9 ubc-14; UBiquitin Conjugating enzyme family member
(ubc-14); K04555 ubiquitin-conjugating enzyme E2 G2 [EC:6.3.2.19]
Length=170
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 57/130 (43%), Gaps = 18/130 (13%)
Query 43 AVDRLRREWKSLQRESLPNVEARP-QEDDFLTWIFLIHDLPEDSSFAGGFYMGKVVFPAT 101
A+ RL E+K L + A P ED+F W LI PE++ FA G + ++ FP
Sbjct 5 ALKRLMTEYKELTTRPPEGIIAAPIDEDNFFEWECLITG-PEETCFANGVFPARITFPQD 63
Query 102 YPFSPPSIYMLTPNGRFETN----RSLCVSFTH----------YHPELWNPSWKVETLLT 147
YP SPP + G F N +C+S H E W+P +E +L
Sbjct 64 YPLSPPKMRFTC--GIFHPNIYADGRVCISILHAPGDDPTGYELSNERWSPVQSIEKILL 121
Query 148 GFVSFMLDEN 157
VS + + N
Sbjct 122 SVVSMLAEPN 131
Lambda K H
0.314 0.127 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40