bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3076_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
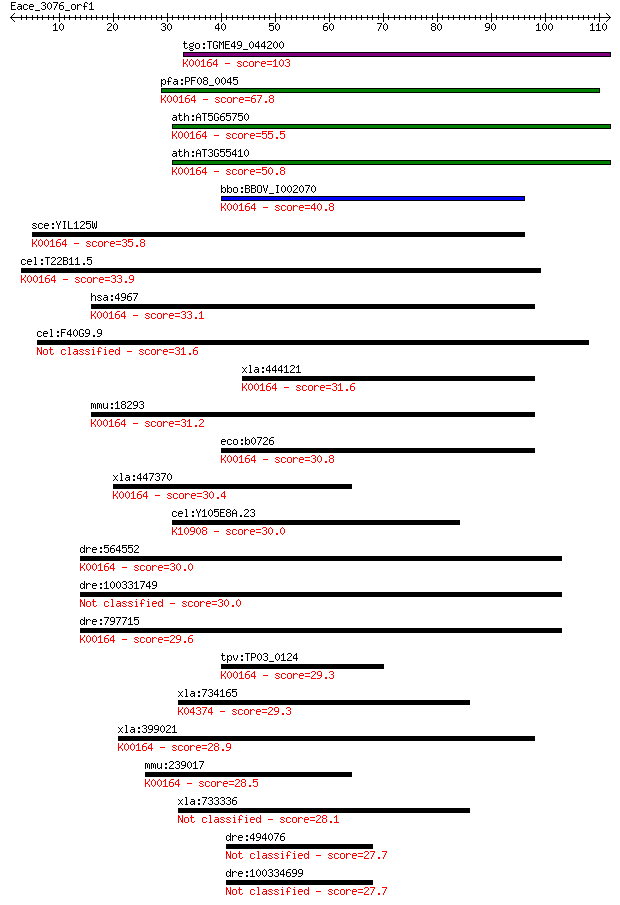
tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1... 103 1e-22
pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.... 67.8 8e-12
ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putat... 55.5 4e-08
ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putat... 50.8 9e-07
bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1 co... 40.8 0.001
sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-k... 35.8 0.032
cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehyd... 33.9 0.14
hsa:4967 OGDH, AKGDH, E1k, OGDC; oxoglutarate (alpha-ketogluta... 33.1 0.25
cel:F40G9.9 hypothetical protein 31.6 0.58
xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogena... 31.6 0.73
mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA41... 31.2 0.87
eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxy... 30.8 1.1
xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like; K... 30.4 1.5
cel:Y105E8A.23 hypothetical protein; K10908 DNA-directed RNA p... 30.0 1.8
dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,... 30.0 1.8
dre:100331749 oxoglutarate (alpha-ketoglutarate) dehydrogenase... 30.0 1.9
dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogen... 29.6 2.2
tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.... 29.3 2.9
xla:734165 atf4-b, MGC196486, MGC196490, atf-4, atf4-ii, creb-... 29.3 3.3
xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alp... 28.9 4.4
mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-)... 28.5 6.2
xla:733336 atf4-iii; Activating Transcription Factor 4 -III 28.1 6.7
dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketol... 27.7 9.5
dre:100334699 probable 2-oxoglutarate dehydrogenase E1 compone... 27.7 9.6
> tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1116
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 53/80 (66%), Positives = 59/80 (73%), Gaps = 1/80 (1%)
Query 33 QSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPL-QLGPEHYGFTAA 91
QSVHDTSRLIQMVRGYQM GHE+AA+NPLSLP++ P+ S P L E YGFT A
Sbjct 187 QSVHDTSRLIQMVRGYQMRGHEIAAVNPLSLPQETPFVSGSRGPTPAGTLDFEAYGFTKA 246
Query 92 DFDKVYVARVPGMQGFLSPD 111
D DKVY RV GM GFLSP+
Sbjct 247 DLDKVYDCRVDGMCGFLSPE 266
> pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1038
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 29/81 (35%), Positives = 49/81 (60%), Gaps = 0/81 (0%)
Query 29 EGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGPEHYGF 88
+G +++++D +R++Q++R YQ GH A INPL LP++PPY S+ ++ +GF
Sbjct 115 KGKTENIYDLARIVQLIRWYQKKGHLYANINPLPLPKEPPYSSVCYEPCKRKMSYVDFGF 174
Query 89 TAADFDKVYVARVPGMQGFLS 109
D DK + +P + GF S
Sbjct 175 NEDDLDKEFFFDLPSISGFSS 195
> ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1025
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 30/81 (37%), Positives = 45/81 (55%), Gaps = 9/81 (11%)
Query 31 SSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGPEHYGFTA 90
S Q++ ++ RL+ +VR YQ+ GH A ++PL L ++ + P L P YGFT
Sbjct 114 SGQTIQESMRLLLLVRAYQVNGHMKAKLDPLGLEKR---------EIPEDLTPGLYGFTE 164
Query 91 ADFDKVYVARVPGMQGFLSPD 111
AD D+ + V M GFLS +
Sbjct 165 ADLDREFFLGVWRMSGFLSEN 185
> ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1017
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 9/81 (11%)
Query 31 SSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGPEHYGFTA 90
S Q++ ++ RL+ +VR YQ+ GH A ++PL L ++ + P L YGFT
Sbjct 111 SGQTIQESMRLLLLVRAYQVNGHMKAKLDPLGLEQR---------EIPEDLDLALYGFTE 161
Query 91 ADFDKVYVARVPGMQGFLSPD 111
AD D+ + V M GF+S +
Sbjct 162 ADLDREFFLGVWQMSGFMSEN 182
> bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1
component (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=891
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 40 RLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGPEHYGFTAADFDK 95
RL ++VR Y+ GH ++ ++PL LPR+PP+ + +L YG D +
Sbjct 23 RLSELVRAYRTEGHCVSTLDPLDLPREPPFHRFIPSDVSTKLCHTTYGLKDEDLGR 78
> sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-ketoglutarate
dehydrogenase complex, which catalyzes a key
step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation
of alpha-ketoglutarate to form succinyl-CoA (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1014
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 42/91 (46%), Gaps = 14/91 (15%)
Query 5 PAATTAGTTGAAAAGSGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLP 64
P T A G A GS + + S+H +L + R YQ+ GH A I+PL +
Sbjct 99 PQGTEAAPLGTAMTGSVD--------ENVSIHLKVQL--LCRAYQVRGHLKAHIDPLGI- 147
Query 65 RKPPYCSLRAAQQPLQLGPEHYGFTAADFDK 95
+ S + P +L ++YGF+ D DK
Sbjct 148 ---SFGSNKNNPVPPELTLDYYGFSKHDLDK 175
> cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1029
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 41/96 (42%), Gaps = 11/96 (11%)
Query 3 ASPAATTAGTTGAAAAGSGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLS 62
SPAA T+ A A S QS+ D ++ ++R YQ GH +A ++PL
Sbjct 109 VSPAAAQVTTSSAPATRLDTN------ASVQSISDHLKIQLLIRSYQTRGHNIADLDPLG 162
Query 63 LPRKPPYCSLRAAQQPLQLGPEHYGFTAADFDKVYV 98
+ ++ P +L YG D D+ ++
Sbjct 163 INSADLDDTI-----PPELELSFYGLGERDLDREFL 193
> hsa:4967 OGDH, AKGDH, E1k, OGDC; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=427
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 38/84 (45%), Gaps = 4/84 (4%)
Query 16 AAAGSGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRA- 74
AA + L A + V D + ++R YQ+ GH +A ++PL + S+ A
Sbjct 105 AAVAHAQSLVEAQPNVDKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGILDADLDSSVPAD 164
Query 75 -AQQPLQLGPEHYGFTAADFDKVY 97
+LG YG +D DKV+
Sbjct 165 IISSTDKLG--FYGLDESDLDKVF 186
> cel:F40G9.9 hypothetical protein
Length=862
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 33/116 (28%), Positives = 45/116 (38%), Gaps = 20/116 (17%)
Query 6 AATTAGTTGAAAAGSGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLP- 64
A G G+ G +S ++GS S SR V G+Q G + I +P
Sbjct 452 GARVPGIQGSRFPGIQGARFSGFQGSRDSEFQGSR----VPGFQ--GARVPGIQGFRVPG 505
Query 65 ----RKPPYCSLRA--AQQPLQLGPEHYG-------FTAADFDKVYVARVPGMQGF 107
R P + R ++ P GP G F +V V+RVPG QGF
Sbjct 506 FQGSRVPGFQGFRVQVSRVPGFQGPGFQGSRVSGSRFPGFQGFRVQVSRVPGFQGF 561
> xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1018
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 30/57 (52%), Gaps = 9/57 (15%)
Query 44 MVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGPEH---YGFTAADFDKVY 97
++R YQ+ GH +A ++PL + S+ P+ +G + YG +D DKV+
Sbjct 132 LIRAYQVRGHHIAKLDPLGIS------SVNFDGAPVIVGSPNVGFYGLEESDLDKVF 182
> mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA4192,
d1401, mKIAA4192; oxoglutarate dehydrogenase (lipoamide)
(EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1023
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 37/84 (44%), Gaps = 4/84 (4%)
Query 16 AAAGSGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRA- 74
A + L A + V D + ++R YQ+ GH +A ++PL + S+ A
Sbjct 105 ATMAHAQSLVEAQPNVDKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGILDADLDSSVPAD 164
Query 75 -AQQPLQLGPEHYGFTAADFDKVY 97
+LG YG +D DKV+
Sbjct 165 IISSTDKLG--FYGLHESDLDKVF 186
> eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxylase,
thiamin-requiring (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=933
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 28/58 (48%), Gaps = 9/58 (15%)
Query 40 RLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGPEHYGFTAADFDKVY 97
+++Q++ Y+ GH+ A ++PL L ++ L P + T ADF + +
Sbjct 91 KVLQLINAYRFRGHQHANLDPLGLWQQDKVAD---------LDPSFHDLTEADFQETF 139
> xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like;
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1018
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 20 SGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSL 63
S +GL +A + + V + + ++R YQ+ GH +A ++PL +
Sbjct 104 SSQGLATAPAKAEKIVEEHLAVQSLIRAYQIRGHHVAQLDPLGI 147
> cel:Y105E8A.23 hypothetical protein; K10908 DNA-directed RNA
polymerase, mitochondrial [EC:2.7.7.6]
Length=1138
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 25/53 (47%), Gaps = 2/53 (3%)
Query 31 SSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQPLQLGP 83
SS ++ D RLI +G + + I PL LP PYC L + L L P
Sbjct 964 SSMALKDWFRLI--AKGSSDLMKTVEWITPLGLPVVQPYCKLVERKGKLILAP 1014
> dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,
zgc:73296; oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1022
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 14/96 (14%)
Query 14 GAAAAG--SGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPL-----SLPRK 66
G + AG + L A + V D + ++R YQ+ GH +A ++PL L
Sbjct 101 GVSLAGLAQAQSLVGAQPNVEKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGIMDADLDSC 160
Query 67 PPYCSLRAAQQPLQLGPEHYGFTAADFDKVYVARVP 102
P + ++ +LG YG +D DKV+ R+P
Sbjct 161 VPTDIITSSD---KLG--FYGLEESDLDKVF--RLP 189
> dre:100331749 oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide)-like
Length=687
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 14/96 (14%)
Query 14 GAAAAG--SGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPL-----SLPRK 66
G + AG + L A + V D + ++R YQ+ GH +A ++PL L
Sbjct 101 GVSLAGLAQAQSLVGAQPNVEKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGIMDADLDSC 160
Query 67 PPYCSLRAAQQPLQLGPEHYGFTAADFDKVYVARVP 102
P + ++ +LG YG +D DKV+ R+P
Sbjct 161 VPTDIITSSD---KLG--FYGLEESDLDKVF--RLP 189
> dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1023
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 12/94 (12%)
Query 14 GAAAAGSGEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPL-----SLPRKPP 68
G + + L A + V D + ++R YQ+ GH +A ++PL L P
Sbjct 104 GLSGLSQSQALIGAQPNVEKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGILDADLDSCVP 163
Query 69 YCSLRAAQQPLQLGPEHYGFTAADFDKVYVARVP 102
+ ++ +LG YG D DKV+ R+P
Sbjct 164 ADIITSSD---KLG--FYGLEETDLDKVF--RLP 190
> tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1030
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 9/30 (30%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 40 RLIQMVRGYQMIGHELAAINPLSLPRKPPY 69
+L ++ Y+ GH ++ ++PL LP++ P+
Sbjct 104 KLNELASAYRTFGHLVSNLDPLKLPKEVPF 133
> xla:734165 atf4-b, MGC196486, MGC196490, atf-4, atf4-ii, creb-2,
creb2, taxreb67, txreb; activating transcription factor
4 (tax-responsive enhancer element B67); K04374 activating
transcription factor 4
Length=342
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 5/59 (8%)
Query 32 SQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQP-----LQLGPEH 85
+ ++ D+ L++ +IGH + + P+SLP P Y A P L LGP+H
Sbjct 108 TSTLEDSCDLLEDPPHPSVIGHFVPSPEPVSLPGSPLYADQVAPVSPDLTESLSLGPDH 166
> xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1021
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 35/79 (44%), Gaps = 4/79 (5%)
Query 21 GEGLYSAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRA--AQQP 78
+ L A + V D + ++R YQ+ GH +A ++PL + + A
Sbjct 109 AQSLLHAQPNVDKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGILDADLDSCVPADIVTSS 168
Query 79 LQLGPEHYGFTAADFDKVY 97
+LG YG +D DKV+
Sbjct 169 DKLG--FYGLQESDLDKVF 185
> mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1029
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 26 SAYEGSSQSVHDTSRLIQMVRGYQMIGHELAAINPLSL 63
S+ +S+ V D + ++R YQ+ GH +A ++PL +
Sbjct 102 SSCTKTSKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGI 139
> xla:733336 atf4-iii; Activating Transcription Factor 4 -III
Length=291
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 5/59 (8%)
Query 32 SQSVHDTSRLIQMVRGYQMIGHELAAINPLSLPRKPPYCSLRAAQQP-----LQLGPEH 85
+ ++ D+ L++ +IGH + + P+SLP P Y A P L LGP+H
Sbjct 108 TSTLEDSCDLLEDPPHPSVIGHFVPSPEPVSLPGSPLYADQVAPVSPDLTESLSLGPDH 166
> dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=925
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 1/27 (3%)
Query 41 LIQMVRGYQMIGHELAAINPLSLPRKP 67
L ++V Y+ GH++A INPL LP+ P
Sbjct 62 LARLVEAYRAHGHKVAKINPL-LPQSP 87
> dre:100334699 probable 2-oxoglutarate dehydrogenase E1 component
DHKTD1, mitochondrial-like
Length=657
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 1/27 (3%)
Query 41 LIQMVRGYQMIGHELAAINPLSLPRKP 67
L ++V Y+ GH++A INPL LP+ P
Sbjct 62 LARLVEAYRAHGHKVAKINPL-LPQSP 87
Lambda K H
0.314 0.130 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2062416360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40