bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3091_orf2
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
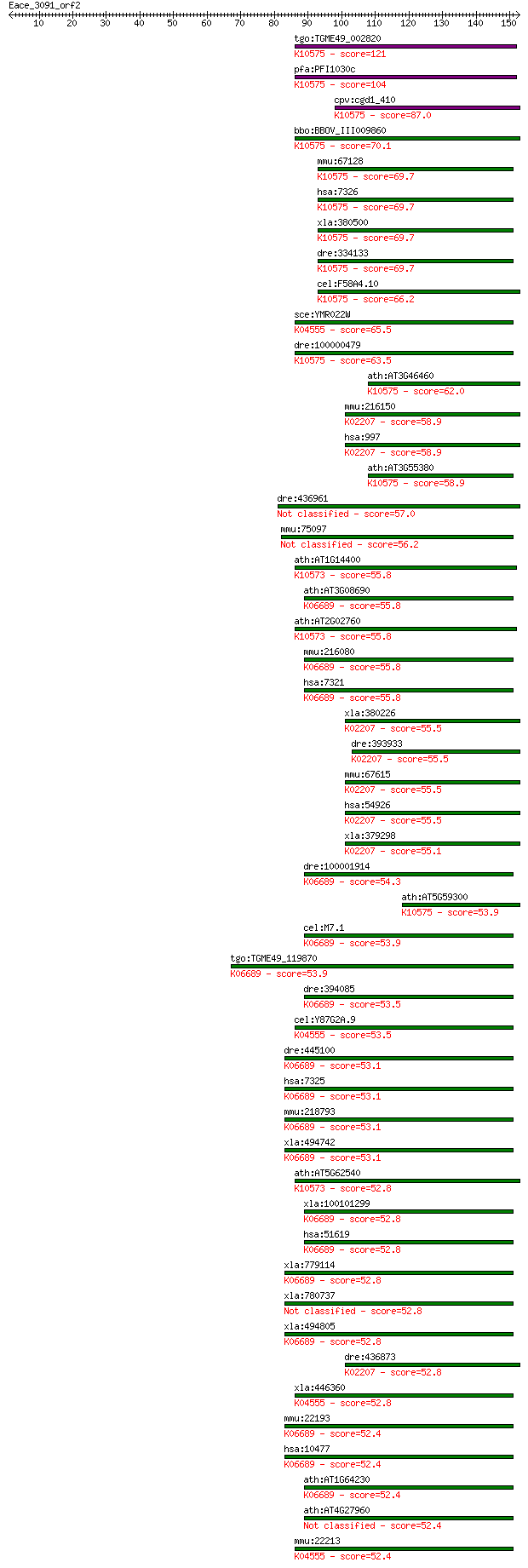
tgo:TGME49_002820 ubiquitin-conjugating enzyme E2, putative (E... 121 7e-28
pfa:PFI1030c ubiquitin conjugating enzyme, putative (EC:6.3.2.... 104 1e-22
cpv:cgd1_410 ubiquitin conjugating enzyme ; K10575 ubiquitin-c... 87.0 2e-17
bbo:BBOV_III009860 17.m07854; ubiquitin conjugating enzyme E2 ... 70.1 3e-12
mmu:67128 Ube2g1, 2700059C12Rik, AI256795, AU014992, AW552068,... 69.7 3e-12
hsa:7326 UBE2G1, E217K, UBC7, UBE2G; ubiquitin-conjugating enz... 69.7 3e-12
xla:380500 ube2g1, MGC53896; ubiquitin-conjugating enzyme E2G ... 69.7 4e-12
dre:334133 ube2g2, UBE2G1, wu:fi33c04, zgc:55974; ubiquitin-co... 69.7 4e-12
cel:F58A4.10 ubc-7; UBiquitin Conjugating enzyme family member... 66.2 4e-11
sce:YMR022W UBC7, QRI8; Ubc7p (EC:6.3.2.19); K04555 ubiquitin-... 65.5 6e-11
dre:100000479 ube2g1b, MGC55321, MGC86866, ube2g1, wu:fc68b05,... 63.5 3e-10
ath:AT3G46460 UBC13; UBC13 (ubiquitin-conjugating enzyme 13); ... 62.0 7e-10
mmu:216150 Cdc34, AI327276, UBE2R1; cell division cycle 34 hom... 58.9 6e-09
hsa:997 CDC34, E2-CDC34, UBC3, UBCH3, UBE2R1; cell division cy... 58.9 6e-09
ath:AT3G55380 UBC14; UBC14 (ubiquitin-conjugating enzyme 14); ... 58.9 7e-09
dre:436961 cdc34b, fb34h09, wu:fb34h09, wu:fe17e09, zgc:91847;... 57.0 2e-08
mmu:75097 4930524E20Rik; RIKEN cDNA 4930524E20 gene 56.2 4e-08
ath:AT1G14400 UBC1; UBC1 (UBIQUITIN CARRIER PROTEIN 1); ubiqui... 55.8 5e-08
ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11); ... 55.8 5e-08
ath:AT2G02760 ATUBC2; ATUBC2 (UBIQUITING-CONJUGATING ENZYME 2)... 55.8 5e-08
mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzy... 55.8 6e-08
hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiqui... 55.8 6e-08
xla:380226 ube2r2, MGC68917; ubiquitin coniugating enzyme 3b; ... 55.5 7e-08
dre:393933 cdc34a, MGC55452, Ube2r2, zgc:55452; cell division ... 55.5 7e-08
mmu:67615 Ube2r2, 1200003M11Rik, Cdc34b, Ubc3b; ubiquitin-conj... 55.5 7e-08
hsa:54926 UBE2R2, CDC34B, FLJ20419, MGC10481, UBC3B; ubiquitin... 55.5 7e-08
xla:379298 cdc34, MGC53533; cell division cycle 34 homolog; K0... 55.1 9e-08
dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjuga... 54.3 2e-07
ath:AT5G59300 UBC7; UBC7 (UBIQUITIN CARRIER PROTEIN 7); protei... 53.9 2e-07
cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquit... 53.9 2e-07
tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containi... 53.9 2e-07
dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689 ub... 53.5 2e-07
cel:Y87G2A.9 ubc-14; UBiquitin Conjugating enzyme family membe... 53.5 3e-07
dre:445100 ube2e2, wu:fb38h07; zgc:92467 (EC:6.3.2.-); K06689 ... 53.1 3e-07
hsa:7325 UBE2E2, FLJ25157, UBCH8; ubiquitin-conjugating enzyme... 53.1 3e-07
mmu:218793 Ube2e2, BC016265, MGC28917; ubiquitin-conjugating e... 53.1 3e-07
xla:494742 ube2e2, ubch8; ubiquitin-conjugating enzyme E2E 2 (... 53.1 4e-07
ath:AT5G62540 UBC3; UBC3 (ubiquitin-conjugating enzyme 3); ubi... 52.8 4e-07
xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating ... 52.8 4e-07
hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzy... 52.8 4e-07
xla:779114 ube2e3-a, MGC81343, ubc15, ubch9, ubcm2, ube2e3, xu... 52.8 4e-07
xla:780737 ubiquitin-conjugating enzyme UBE2E3 52.8 4e-07
xla:494805 hypothetical LOC494805; K06689 ubiquitin-conjugatin... 52.8 4e-07
dre:436873 ube2r2, zgc:92307; ubiquitin-conjugating enzyme E2R... 52.8 5e-07
xla:446360 ube2g2, MGC82328; ubiquitin-conjugating enzyme E2G ... 52.8 5e-07
mmu:22193 Ube2e3, Ubce4, ubcM2; ubiquitin-conjugating enzyme E... 52.4 5e-07
hsa:10477 UBE2E3, UBCH9, UbcM2; ubiquitin-conjugating enzyme E... 52.4 5e-07
ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative; K... 52.4 5e-07
ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubi... 52.4 6e-07
mmu:22213 Ube2g2, 1110003O05Rik, D10Xrf369, UBC7, Ubc7p; ubiqu... 52.4 6e-07
> tgo:TGME49_002820 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10575 ubiquitin-conjugating enzyme E2 G1 [EC:6.3.2.19]
Length=168
Score = 121 bits (304), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 55/68 (80%), Positives = 60/68 (88%), Gaps = 2/68 (2%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDE--GNFFSWRVCLEGPPDTFYEGGIFQASL 143
M+N+ARELLKKQF+ELSRD PSGCSVGLDDE G+FF WRVC EGPPDT YEGGIF A+L
Sbjct 1 MSNIARELLKKQFLELSRDCPSGCSVGLDDEAGGDFFVWRVCFEGPPDTLYEGGIFTAAL 60
Query 144 KFPQDFPN 151
KFP DFPN
Sbjct 61 KFPPDFPN 68
> pfa:PFI1030c ubiquitin conjugating enzyme, putative (EC:6.3.2.19);
K10575 ubiquitin-conjugating enzyme E2 G1 [EC:6.3.2.19]
Length=163
Score = 104 bits (259), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 46/66 (69%), Positives = 52/66 (78%), Gaps = 0/66 (0%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M N+ARELLKKQFIEL+RD +G SVGL DE NFF W VC EGP +T YEGGI+ A+L F
Sbjct 1 MANIARELLKKQFIELTRDHNAGFSVGLVDESNFFEWNVCFEGPKNTLYEGGIYNATLSF 60
Query 146 PQDFPN 151
P DFPN
Sbjct 61 PSDFPN 66
> cpv:cgd1_410 ubiquitin conjugating enzyme ; K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=156
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 39/55 (70%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 98 FIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
+ EL RD SG SVGLDD+ NF WRVC EGPPDT YEGGIF A L FP+DFPNS
Sbjct 4 YSELIRDDTSGFSVGLDDDSNFLRWRVCFEGPPDTLYEGGIFNALLTFPEDFPNS 58
> bbo:BBOV_III009860 17.m07854; ubiquitin conjugating enzyme E2
(EC:6.3.2.19); K10575 ubiquitin-conjugating enzyme E2 G1 [EC:6.3.2.19]
Length=164
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M +ARE+L++Q+ EL+R SVGL+D+ + F WR+C +GP T +EGGIF F
Sbjct 1 MEKIAREILRRQYTELTRSDNCYFSVGLEDD-DLFKWRICFQGPYGTPFEGGIFTVITTF 59
Query 146 PQDFPNS 152
P +FPN
Sbjct 60 PNNFPND 66
> mmu:67128 Ube2g1, 2700059C12Rik, AI256795, AU014992, AW552068,
D130023C12Rik; ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog,
C. elegans) (EC:6.3.2.19); K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=170
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 93 LLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFP 150
LL++Q EL+++ G S GL D+ + + W V + GPPDT YEGG+F+A L FP+D+P
Sbjct 9 LLRRQLAELNKNPVEGFSAGLIDDNDLYRWEVLIIGPPDTLYEGGVFKAHLTFPKDYP 66
> hsa:7326 UBE2G1, E217K, UBC7, UBE2G; ubiquitin-conjugating enzyme
E2G 1 (UBC7 homolog, yeast) (EC:6.3.2.19); K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=170
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 93 LLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFP 150
LL++Q EL+++ G S GL D+ + + W V + GPPDT YEGG+F+A L FP+D+P
Sbjct 9 LLRRQLAELNKNPVEGFSAGLIDDNDLYRWEVLIIGPPDTLYEGGVFKAHLTFPKDYP 66
> xla:380500 ube2g1, MGC53896; ubiquitin-conjugating enzyme E2G
1 (UBC7 homolog) (EC:6.3.2.19); K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=170
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 93 LLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFP 150
LL++Q EL+++ G S GL D+ + + W V + GPPDT YEGG+F+A L FP+D+P
Sbjct 9 LLRRQLAELNKNPVEGFSAGLIDDNDLYRWEVLIIGPPDTLYEGGVFKAHLTFPRDYP 66
> dre:334133 ube2g2, UBE2G1, wu:fi33c04, zgc:55974; ubiquitin-conjugating
enzyme E2G 2 (EC:6.3.2.-); K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=170
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 93 LLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFP 150
LL++Q EL+++ G S GL D+ + + W V + GPPDT YEGG+F+A L FP+D+P
Sbjct 9 LLRRQLAELNKNPVEGFSAGLIDDNDLYRWEVLIIGPPDTLYEGGVFKAHLTFPKDYP 66
> cel:F58A4.10 ubc-7; UBiquitin Conjugating enzyme family member
(ubc-7); K10575 ubiquitin-conjugating enzyme E2 G1 [EC:6.3.2.19]
Length=164
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 93 LLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
LLKKQ ++ R G S GL D+ + + W V + GPPDT YEGG F+A L FP+D+P
Sbjct 7 LLKKQLADMRRVPVDGFSAGLVDDNDIYKWEVLVIGPPDTLYEGGFFKAILDFPRDYPQK 66
> sce:YMR022W UBC7, QRI8; Ubc7p (EC:6.3.2.19); K04555 ubiquitin-conjugating
enzyme E2 G2 [EC:6.3.2.19]
Length=165
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M+ A++ L K+ +L +D+P G G E N F W ++GPPDT Y G+F A L+F
Sbjct 1 MSKTAQKRLLKELQQLIKDSPPGIVAGPKSENNIFIWDCLIQGPPDTPYADGVFNAKLEF 60
Query 146 PQDFP 150
P+D+P
Sbjct 61 PKDYP 65
> dre:100000479 ube2g1b, MGC55321, MGC86866, ube2g1, wu:fc68b05,
wu:fc75g08, wu:fk32c04, zgc:55321; ubiquitin-conjugating
enzyme E2G 1b (UBC7 homolog, yeast) (EC:6.3.2.19); K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=169
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 41/65 (63%), Gaps = 0/65 (0%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
MT + LL+KQ EL+++ G S GL D+ + + W V + GP DT +EGG F+A L F
Sbjct 1 MTEQSALLLRKQLAELNKNPVEGFSAGLIDDDDIYQWEVVVIGPQDTMFEGGFFKAHLIF 60
Query 146 PQDFP 150
P D+P
Sbjct 61 PHDYP 65
> ath:AT3G46460 UBC13; UBC13 (ubiquitin-conjugating enzyme 13);
ubiquitin-protein ligase; K10575 ubiquitin-conjugating enzyme
E2 G1 [EC:6.3.2.19]
Length=166
Score = 62.0 bits (149), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 27/45 (60%), Positives = 31/45 (68%), Gaps = 0/45 (0%)
Query 108 GCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
G S GL DE N F W V + GPPDT YEGG F A + FPQ++PNS
Sbjct 23 GFSAGLVDEKNIFEWSVTIIGPPDTLYEGGFFYAIMSFPQNYPNS 67
> mmu:216150 Cdc34, AI327276, UBE2R1; cell division cycle 34 homolog
(S. cerevisiae) (EC:6.3.2.19); K02207 ubiquitin-conjugating
enzyme E2 R [EC:6.3.2.19]
Length=235
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 26/52 (50%), Positives = 35/52 (67%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G V L DEG+ ++W V + GPP+T+YEGG F+A LKFP D+P S
Sbjct 20 LQEEPVEGFRVTLVDEGDLYNWEVAIFGPPNTYYEGGYFKARLKFPIDYPYS 71
> hsa:997 CDC34, E2-CDC34, UBC3, UBCH3, UBE2R1; cell division
cycle 34 homolog (S. cerevisiae) (EC:6.3.2.19); K02207 ubiquitin-conjugating
enzyme E2 R [EC:6.3.2.19]
Length=236
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 26/52 (50%), Positives = 35/52 (67%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G V L DEG+ ++W V + GPP+T+YEGG F+A LKFP D+P S
Sbjct 20 LQEEPVEGFRVTLVDEGDLYNWEVAIFGPPNTYYEGGYFKARLKFPIDYPYS 71
> ath:AT3G55380 UBC14; UBC14 (ubiquitin-conjugating enzyme 14);
ubiquitin-protein ligase; K10575 ubiquitin-conjugating enzyme
E2 G1 [EC:6.3.2.19]
Length=167
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 108 GCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFP 150
G S GL DE N F W V + GPPDT YEGG F A + FP+++P
Sbjct 24 GFSAGLVDEKNVFQWSVSIMGPPDTLYEGGFFNAIMSFPENYP 66
> dre:436961 cdc34b, fb34h09, wu:fb34h09, wu:fe17e09, zgc:91847;
cell division cycle 34 homolog (S. cerevisiae) b (EC:6.3.2.-)
Length=239
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 81 RDSAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQ 140
+ S+A +++ L + L + G + L DE + ++W V + GPP+T YEGG F+
Sbjct 3 QQSSAQVASSQKALMLEMKSLQEEPVEGFKITLVDEADLYNWEVAIFGPPNTHYEGGYFK 62
Query 141 ASLKFPQDFPNS 152
A +KFP D+P S
Sbjct 63 ARIKFPVDYPYS 74
> mmu:75097 4930524E20Rik; RIKEN cDNA 4930524E20 gene
Length=155
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 43/69 (62%), Gaps = 1/69 (1%)
Query 82 DSAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQA 141
D + + +A + ++K+ + +S+D P+ CS G E N F W+ + GP D+ Y+GG+F
Sbjct 2 DQSTLGAMALKRIQKELVAISQDPPAHCSAGPVAE-NMFHWQATIMGPEDSPYQGGVFFL 60
Query 142 SLKFPQDFP 150
S+ FP ++P
Sbjct 61 SVHFPNNYP 69
> ath:AT1G14400 UBC1; UBC1 (UBIQUITIN CARRIER PROTEIN 1); ubiquitin-protein
ligase; K10573 ubiquitin-conjugating enzyme E2
A [EC:6.3.2.19]
Length=152
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M+ AR+ L + F L +D P+G S G + N W + GP DT ++GG F+ SL+F
Sbjct 1 MSTPARKRLMRDFKRLQQDPPAGIS-GAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQF 59
Query 146 PQDFPN 151
+D+PN
Sbjct 60 SEDYPN 65
> ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + + K+ +L +D PS CS G E + F W+ + GPP++ Y GG+F S+ FP D
Sbjct 1 MASKRILKELKDLQKDPPSNCSAGPVAE-DMFHWQATIMGPPESPYAGGVFLVSIHFPPD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> ath:AT2G02760 ATUBC2; ATUBC2 (UBIQUITING-CONJUGATING ENZYME
2); ubiquitin-protein ligase; K10573 ubiquitin-conjugating enzyme
E2 A [EC:6.3.2.19]
Length=152
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M+ AR+ L + F L +D P+G S G + N W + GP DT ++GG F+ SL+F
Sbjct 1 MSTPARKRLMRDFKRLQQDPPAGIS-GAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQF 59
Query 146 PQDFPN 151
+D+PN
Sbjct 60 SEDYPN 65
> mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzyme
E2D 1, UBC4/5 homolog (yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ +L RD P+ CS G + + F W+ + GPPD+ Y+GG+F ++ FP D
Sbjct 1 MALKRIQKELSDLQRDPPAHCSAGPVGD-DLFHWQATIMGPPDSAYQGGVFFLTVHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ +L RD P+ CS G + + F W+ + GPPD+ Y+GG+F ++ FP D
Sbjct 1 MALKRIQKELSDLQRDPPAHCSAGPVGD-DLFHWQATIMGPPDSAYQGGVFFLTVHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> xla:380226 ube2r2, MGC68917; ubiquitin coniugating enzyme 3b;
K02207 ubiquitin-conjugating enzyme E2 R [EC:6.3.2.19]
Length=238
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G + L DE + ++W V + GPP+T YEGG F+A +KFP D+P S
Sbjct 20 LQEEPVEGFRITLVDESDLYNWEVAIFGPPNTLYEGGYFKAHIKFPIDYPYS 71
> dre:393933 cdc34a, MGC55452, Ube2r2, zgc:55452; cell division
cycle 34 homolog (S. cerevisiae) a (EC:6.3.2.-); K02207 ubiquitin-conjugating
enzyme E2 R [EC:6.3.2.19]
Length=241
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/51 (47%), Positives = 34/51 (66%), Gaps = 1/51 (1%)
Query 103 RDAP-SGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
+D P G + L DE + ++W V + GPP+T YEGG F+A +KFP D+P S
Sbjct 24 QDEPVEGFKITLVDESDLYNWEVAIFGPPNTHYEGGYFKARIKFPIDYPYS 74
> mmu:67615 Ube2r2, 1200003M11Rik, Cdc34b, Ubc3b; ubiquitin-conjugating
enzyme E2R 2 (EC:6.3.2.19); K02207 ubiquitin-conjugating
enzyme E2 R [EC:6.3.2.19]
Length=238
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G + L DE + ++W V + GPP+T YEGG F+A +KFP D+P S
Sbjct 20 LQEEPVEGFRITLVDESDLYNWEVAIFGPPNTLYEGGYFKAHIKFPIDYPYS 71
> hsa:54926 UBE2R2, CDC34B, FLJ20419, MGC10481, UBC3B; ubiquitin-conjugating
enzyme E2R 2 (EC:6.3.2.19); K02207 ubiquitin-conjugating
enzyme E2 R [EC:6.3.2.19]
Length=238
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G + L DE + ++W V + GPP+T YEGG F+A +KFP D+P S
Sbjct 20 LQEEPVEGFRITLVDESDLYNWEVAIFGPPNTLYEGGYFKAHIKFPIDYPYS 71
> xla:379298 cdc34, MGC53533; cell division cycle 34 homolog;
K02207 ubiquitin-conjugating enzyme E2 R [EC:6.3.2.19]
Length=237
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 25/52 (48%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G V L EG+ ++W V + GPP+T YEGG F+A LKFP D+P S
Sbjct 22 LQEEPVEGFRVTLVGEGDLYNWEVAIFGPPNTLYEGGYFKARLKFPVDYPYS 73
> dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ +L RD P+ CS G E + F W+ + GP D+ Y+GG+F ++ FP D
Sbjct 1 MALKRIQKELTDLQRDPPAQCSAGPVGE-DLFHWQATIMGPNDSPYQGGVFFLTIHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> ath:AT5G59300 UBC7; UBC7 (UBIQUITIN CARRIER PROTEIN 7); protein
binding / ubiquitin-protein ligase; K10575 ubiquitin-conjugating
enzyme E2 G1 [EC:6.3.2.19]
Length=198
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 118 NFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
N F W V + GPPDT YEGG F A + FPQ++PNS
Sbjct 65 NIFEWSVTIIGPPDTLYEGGFFNAIMTFPQNYPNS 99
> cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ +L RD P+ CS G + + F W+ + GPP++ Y+GG+F ++ FP D
Sbjct 1 MALKRIQKELQDLGRDPPAQCSAGPVGD-DLFHWQATIMGPPESPYQGGVFFLTIHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containing
protein (EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=345
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 45/84 (53%), Gaps = 1/84 (1%)
Query 67 AAVISCTCFLLFVLRDSAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCL 126
A V + L + ++ +A + + K+ +LS+D P+ CS G + + F W+ +
Sbjct 177 AKVHPASSPRLRSIHTDTEVSTMALKRINKELNDLSKDPPTNCSAGPVGD-DMFHWQATI 235
Query 127 EGPPDTFYEGGIFQASLKFPQDFP 150
GP D+ Y GG+F ++ FP D+P
Sbjct 236 MGPEDSPYSGGVFFLNIHFPSDYP 259
> dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ +L RD PS CS G E + F W+ + GP D+ Y+GG+F ++ FP D
Sbjct 1 MALKRIQKELQDLQRDPPSQCSAGPLGE-DLFHWQATIMGPGDSPYQGGVFFLTIHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> cel:Y87G2A.9 ubc-14; UBiquitin Conjugating enzyme family member
(ubc-14); K04555 ubiquitin-conjugating enzyme E2 G2 [EC:6.3.2.19]
Length=170
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M A + L ++ EL+ P G DE NFF W + GP +T + G+F A + F
Sbjct 1 MAGYALKRLMTEYKELTTRPPEGIIAAPIDEDNFFEWECLITGPEETCFANGVFPARITF 60
Query 146 PQDFP 150
PQD+P
Sbjct 61 PQDYP 65
> dre:445100 ube2e2, wu:fb38h07; zgc:92467 (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+AA + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 49 TAAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 107
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 108 IAFTPDYP 115
> hsa:7325 UBE2E2, FLJ25157, UBCH8; ubiquitin-conjugating enzyme
E2E 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+AA + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 49 TAAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 107
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 108 ITFSPDYP 115
> mmu:218793 Ube2e2, BC016265, MGC28917; ubiquitin-conjugating
enzyme E2E 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+AA + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 49 TAAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 107
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 108 ITFSPDYP 115
> xla:494742 ube2e2, ubch8; ubiquitin-conjugating enzyme E2E 2
(UBC4/5 homolog); K06689 ubiquitin-conjugating enzyme E2 D/E
[EC:6.3.2.19]
Length=201
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+AA + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 49 TAAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 107
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 108 ITFSPDYP 115
> ath:AT5G62540 UBC3; UBC3 (ubiquitin-conjugating enzyme 3); ubiquitin-protein
ligase; K10573 ubiquitin-conjugating enzyme
E2 A [EC:6.3.2.19]
Length=150
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
MT A++ L F L +D P G S G + N W + GP DT ++GG F+ +L F
Sbjct 1 MTTPAKKRLMWDFKRLQKDPPVGIS-GAPQDNNIMHWNALIFGPEDTPWDGGTFKLTLHF 59
Query 146 PQDFPNS 152
+D+PN
Sbjct 60 TEDYPNK 66
> xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ ++L RD P+ CS G E + F W+ + GP D+ ++GG+F ++ FP D
Sbjct 1 MALKRIQKELMDLQRDPPAQCSAGPVGE-DLFHWQATIMGPNDSPFQGGVFFLTIHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzyme
E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + ++K+ +L RD P+ CS G + + F W+ + GP D+ Y+GG+F ++ FP D
Sbjct 1 MALKRIQKELTDLQRDPPAQCSAGPVGD-DLFHWQATIMGPNDSPYQGGVFFLTIHFPTD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> xla:779114 ube2e3-a, MGC81343, ubc15, ubch9, ubcm2, ube2e3,
xubc15; ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog)
(EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=207
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+ A + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 55 TTAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 113
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 114 ITFSSDYP 121
> xla:780737 ubiquitin-conjugating enzyme UBE2E3
Length=207
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+ A + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 55 TTAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 113
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 114 ITFSSDYP 121
> xla:494805 hypothetical LOC494805; K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+AA + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 49 TAAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 107
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 108 IAFSPDYP 115
> dre:436873 ube2r2, zgc:92307; ubiquitin-conjugating enzyme E2R
2 (EC:6.3.2.-); K02207 ubiquitin-conjugating enzyme E2 R
[EC:6.3.2.19]
Length=250
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 101 LSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQDFPNS 152
L + G + L +E + ++W V + GPP+T YEGG F+A +KFP D+P S
Sbjct 20 LQEEPVEGFRITLVEESDLYNWEVAIFGPPNTLYEGGYFKAHIKFPIDYPYS 71
> xla:446360 ube2g2, MGC82328; ubiquitin-conjugating enzyme E2G
2 (UBC7 homolog); K04555 ubiquitin-conjugating enzyme E2 G2
[EC:6.3.2.19]
Length=165
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M A + L ++ +L+ + P G G +E NFF W + GP DT +E G+F A L F
Sbjct 1 MAGTALKRLMAEYKQLTLNPPEGIVAGPINEENFFEWEALIMGPEDTCFECGVFPAILSF 60
Query 146 PQDFP 150
P D+P
Sbjct 61 PLDYP 65
> mmu:22193 Ube2e3, Ubce4, ubcM2; ubiquitin-conjugating enzyme
E2E 3, UBC4/5 homolog (yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=207
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+ A + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 55 TTAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 113
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 114 ITFSSDYP 121
> hsa:10477 UBE2E3, UBCH9, UbcM2; ubiquitin-conjugating enzyme
E2E 3 (UBC4/5 homolog, yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=207
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 83 SAAMTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQAS 142
+ A + + + ++K+ E++ D P CS G + N + WR + GPP + YEGG+F
Sbjct 55 TTAKLSTSAKRIQKELAEITLDPPPNCSAGPKGD-NIYEWRSTILGPPGSVYEGGVFFLD 113
Query 143 LKFPQDFP 150
+ F D+P
Sbjct 114 ITFSSDYP 121
> ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + + K+ +L +D P+ CS G E + F W+ + GP D+ Y GG+F ++ FP D
Sbjct 1 MASKRILKELKDLQKDPPTSCSAGPVAE-DMFHWQATIMGPSDSPYSGGVFLVTIHFPPD 59
Query 149 FP 150
+P
Sbjct 60 YP 61
> ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubiquitin-protein
ligase
Length=178
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 89 VARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKFPQD 148
+A + + K+ +L +D P+ CS G E + F W+ + GP D+ Y GG+F ++ FP D
Sbjct 31 MASKRILKELKDLQKDPPTSCSAGPVAE-DMFHWQATIMGPSDSPYSGGVFLVTIHFPPD 89
Query 149 FP 150
+P
Sbjct 90 YP 91
> mmu:22213 Ube2g2, 1110003O05Rik, D10Xrf369, UBC7, Ubc7p; ubiquitin-conjugating
enzyme E2G 2 (EC:6.3.2.19); K04555 ubiquitin-conjugating
enzyme E2 G2 [EC:6.3.2.19]
Length=165
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 86 MTNVARELLKKQFIELSRDAPSGCSVGLDDEGNFFSWRVCLEGPPDTFYEGGIFQASLKF 145
M A + L ++ +L+ + P G G +E NFF W + GP DT +E G+F A L F
Sbjct 1 MAGTALKRLMAEYKQLTLNPPEGIVAGPMNEENFFEWEALIMGPEDTCFEFGVFPAILSF 60
Query 146 PQDFP 150
P D+P
Sbjct 61 PLDYP 65
Lambda K H
0.329 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40