bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3099_orf1
Length=68
Score E
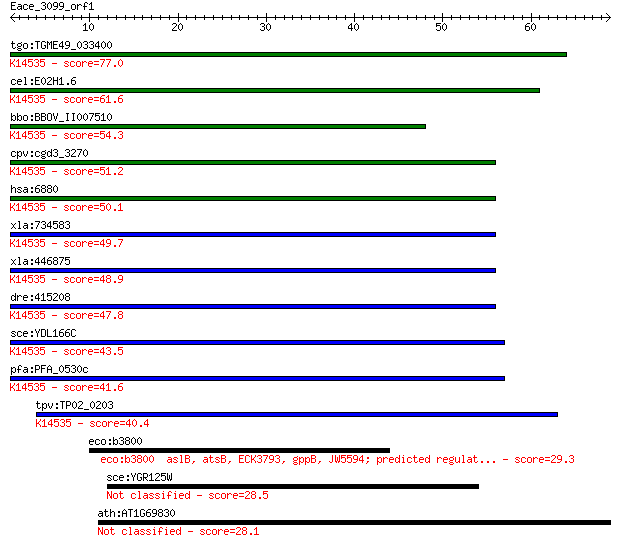
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033400 UPF0101 protein CGI-137, putative (EC:2.7.4.... 77.0 2e-14
cel:E02H1.6 hypothetical protein; K14535 transcription initiat... 61.6 6e-10
bbo:BBOV_II007510 18.m06623; hypothetical protein; K14535 tran... 54.3 1e-07
cpv:cgd3_3270 possible nucleotide kinase related to CMP and AM... 51.2 7e-07
hsa:6880 TAF9, AK6, CGI-137, CINAP, CIP, MGC1603, MGC3647, MGC... 50.1 2e-06
xla:734583 taf9, MGC115514; TAF9 RNA polymerase II, TATA box b... 49.7 2e-06
xla:446875 MGC80885 protein; K14535 transcription initiation f... 48.9 4e-06
dre:415208 zgc:86811; K14535 transcription initiation factor T... 47.8 9e-06
sce:YDL166C FAP7; Essential NTPase required for small ribosome... 43.5 2e-04
pfa:PFA_0530c adenylate kinase, putative; K14535 transcription... 41.6 6e-04
tpv:TP02_0203 hypothetical protein; K14535 transcription initi... 40.4 0.001
eco:b3800 aslB, atsB, ECK3793, gppB, JW5594; predicted regulat... 29.3 2.8
sce:YGR125W Putative protein of unknown function; deletion mut... 28.5 4.9
ath:AT1G69830 AMY3; AMY3 (ALPHA-AMYLASE-LIKE 3); alpha-amylase... 28.1 6.9
> tgo:TGME49_033400 UPF0101 protein CGI-137, putative (EC:2.7.4.3);
K14535 transcription initiation factor TFIID subunit 9
/ adenylate kinase [EC:2.7.4.3]
Length=170
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/63 (57%), Positives = 47/63 (74%), Gaps = 0/63 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNVTAAAE 60
L+DRL R Y ++KI EN++ EIF+VLLDEA E FGEDK+KELQ+N L+DNV+A
Sbjct 103 LYDRLEKRHYKEEKIRENVECEIFRVLLDEAIEAFGEDKVKELQSNNFTDLSDNVSAVMT 162
Query 61 AYN 63
+ N
Sbjct 163 SVN 165
> cel:E02H1.6 hypothetical protein; K14535 transcription initiation
factor TFIID subunit 9 / adenylate kinase [EC:2.7.4.3]
Length=182
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 42/60 (70%), Gaps = 0/60 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNVTAAAE 60
L+DRL +R Y + KI EN++ EIF LL+EARE++ ED + ELQ+ T +Q+ +N+ E
Sbjct 110 LYDRLQSRGYSEFKIKENVECEIFGTLLEEARESYSEDIVHELQSETTEQMEENLERICE 169
> bbo:BBOV_II007510 18.m06623; hypothetical protein; K14535 transcription
initiation factor TFIID subunit 9 / adenylate kinase
[EC:2.7.4.3]
Length=172
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/47 (55%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNT 47
L RL+ R Y KKID N++AEIF+V L +A + FGEDK+ EL +NT
Sbjct 105 LSKRLSDRGYGDKKIDCNIEAEIFKVCLQDAVDHFGEDKVVELPSNT 151
> cpv:cgd3_3270 possible nucleotide kinase related to CMP and
AMP kinases ; K14535 transcription initiation factor TFIID subunit
9 / adenylate kinase [EC:2.7.4.3]
Length=179
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNV 55
LFDRL R Y Q KI EN++ EIF+V+ +A E F E++I +LQ+N I + N+
Sbjct 112 LFDRLEKRGYSQDKIKENVECEIFKVIRFDATEIFDEEQIIDLQSNNICEQRSNI 166
> hsa:6880 TAF9, AK6, CGI-137, CINAP, CIP, MGC1603, MGC3647, MGC5067,
MGC:1603, MGC:3647, MGC:5067, TAF2G, TAFII31, TAFII32,
TAFIID32, hCINAP; TAF9 RNA polymerase II, TATA box binding
protein (TBP)-associated factor, 32kDa (EC:2.7.4.3); K14535
transcription initiation factor TFIID subunit 9 / adenylate
kinase [EC:2.7.4.3]
Length=169
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNV 55
L++RL R Y +KK+ +N+Q EIFQVL +EA ++ E+ + +L +N ++L +NV
Sbjct 99 LYERLETRGYNEKKLTDNIQCEIFQVLYEEATASYKEEIVHQLPSNKPEELENNV 153
> xla:734583 taf9, MGC115514; TAF9 RNA polymerase II, TATA box
binding protein (TBP)-associated factor, 32kDa; K14535 transcription
initiation factor TFIID subunit 9 / adenylate kinase
[EC:2.7.4.3]
Length=171
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 38/55 (69%), Gaps = 0/55 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNV 55
L+ RL +R Y +KK+ +N+Q EIFQ + +EA E++ +D + +L +NT + L N+
Sbjct 102 LYQRLESRGYKEKKLQDNIQCEIFQTIYEEAAESYQKDIVHQLPSNTPEDLEQNI 156
> xla:446875 MGC80885 protein; K14535 transcription initiation
factor TFIID subunit 9 / adenylate kinase [EC:2.7.4.3]
Length=171
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNV 55
L++RL +R Y +KK+ +N+Q EIFQ + +EA E++ ++ + +L +NT + L N+
Sbjct 102 LYERLESRGYKEKKLQDNIQCEIFQTIYEEAAESYQKEIVHQLPSNTPEDLEQNI 156
> dre:415208 zgc:86811; K14535 transcription initiation factor
TFIID subunit 9 / adenylate kinase [EC:2.7.4.3]
Length=171
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNV 55
L++RL +R Y KK+ +N+Q EIFQ + +EA E + E+ + +L +NT + L N+
Sbjct 102 LYNRLESRGYTGKKLQDNVQCEIFQTIFEEAVEAYKEEIVHQLPSNTPEDLERNL 156
> sce:YDL166C FAP7; Essential NTPase required for small ribosome
subunit synthesis, mediates processing of the 20S pre-rRNA
at site D in the cytoplasm but associates only transiently
with 43S preribosomes via Rps14p, may be the endonuclease for
site D; K14535 transcription initiation factor TFIID subunit
9 / adenylate kinase [EC:2.7.4.3]
Length=197
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 35/56 (62%), Gaps = 0/56 (0%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNVT 56
L+ RL AR Y KI+EN+ AEI V+ +A E++ + ELQ++T + + NV+
Sbjct 107 LYSRLHARGYHDSKIEENLDAEIMGVVKQDAVESYEPHIVVELQSDTKEDMVSNVS 162
> pfa:PFA_0530c adenylate kinase, putative; K14535 transcription
initiation factor TFIID subunit 9 / adenylate kinase [EC:2.7.4.3]
Length=186
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Query 1 LFDRLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKI-KELQNNTIQQLNDNVT 56
L++RL R Y ++KI N++ EIFQV+ ++ + F I +E++NN +QQ ++N++
Sbjct 120 LYERLEKRNYTKEKIKNNIECEIFQVIKEDILDHFPNTNILQEIENNDLQQYDNNLS 176
> tpv:TP02_0203 hypothetical protein; K14535 transcription initiation
factor TFIID subunit 9 / adenylate kinase [EC:2.7.4.3]
Length=184
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 4 RLTARQYPQKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLNDNVTAAAEAY 62
RL AR Y + K+ +N+Q EIFQ L ++ E F K+ + +N + L +N + + +
Sbjct 121 RLEARNYSESKVKQNLQCEIFQTCLFDSYEVFDRYKVIPVDSNNEEDLENNTQSVIDYF 179
> eco:b3800 aslB, atsB, ECK3793, gppB, JW5594; predicted regulator
of arylsulfatase activity; K06871
Length=411
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 10 YPQKKIDENMQAEIFQVLLDEARETFGEDKIKEL 43
YPQ ++ Q I +++ ++ FGEDK K+L
Sbjct 302 YPQYRLGNMHQQTIAEMIDSPQQQAFGEDKFKQL 335
> sce:YGR125W Putative protein of unknown function; deletion mutant
has decreased rapamycin resistance but normal wormannin
resistance; green fluorescent protein (GFP)-fusion protein
localizes to the vacuole
Length=1036
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 12 QKKIDENMQAEIFQVLLDEARETFGEDKIKELQNNTIQQLND 53
++ IDE E ++LL A +D+ + LQN+++ +L D
Sbjct 132 EENIDEEYSDEYSRLLLSPASSNVDDDRNRGLQNSSLPELED 173
> ath:AT1G69830 AMY3; AMY3 (ALPHA-AMYLASE-LIKE 3); alpha-amylase
(EC:3.2.1.1)
Length=887
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 35/67 (52%), Gaps = 13/67 (19%)
Query 11 PQKKIDENMQA---------EIFQVLLDEARETFGEDKIKELQNNTIQQLNDNVTAAAEA 61
P++K D+ + A EI + +D + + +KE+Q N +Q++ AAEA
Sbjct 403 PKRKTDKEVSASGFTKEIITEIRNLAIDISSHKNQKTNVKEVQENILQEIE---KLAAEA 459
Query 62 YNNLFRS 68
Y ++FRS
Sbjct 460 Y-SIFRS 465
Lambda K H
0.315 0.130 0.344
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033830404
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40