bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3134_orf2
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
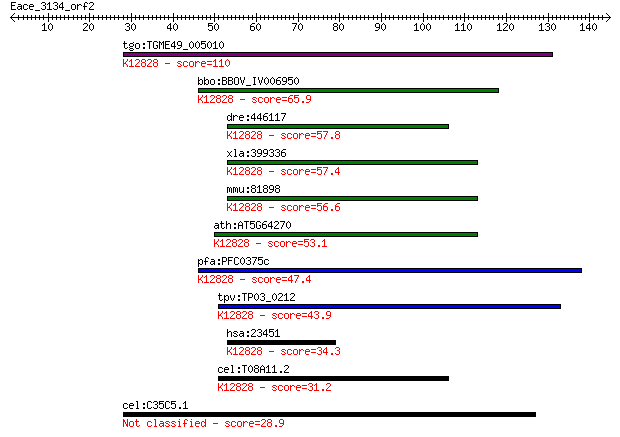
tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5... 110 1e-24
bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing ... 65.9 4e-11
dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1; K... 57.8 1e-08
xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155; sp... 57.4 2e-08
mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP15... 56.6 3e-08
ath:AT5G64270 splicing factor, putative; K12828 splicing facto... 53.1 3e-07
pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828 sp... 47.4 1e-05
tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing fa... 43.9 2e-04
hsa:23451 SF3B1, Hsh155, PRP10, PRPF10, SAP155, SF3b155; splic... 34.3 0.12
cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B s... 31.2 1.1
cel:C35C5.1 sdc-2; Sex determination and Dosage Compensation d... 28.9 6.0
> tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5.5.1.4);
K12828 splicing factor 3B subunit 1
Length=1386
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 65/106 (61%), Positives = 78/106 (73%), Gaps = 6/106 (5%)
Query 28 SERRAMLASSGGDAGDAAALLERRAIAAREDMYRRQRFQRALSPERQDPFAADGGRRADV 87
+ER ML + GD +A + ++IAARED YRRQRFQRALSPER DPF+ DG A
Sbjct 78 AERERMLMAQQGDGVEATGV--DKSIAAREDFYRRQRFQRALSPERADPFS-DGDLGAAS 134
Query 88 ---SARSYADVMVEQQLDREKQAAVKQIRKLQEDAAIAQQLLQQRQ 130
SAR+YADVM+EQ LDREKQAAV+QIRKLQEDA + +QL Q+Q
Sbjct 135 RGESARTYADVMLEQHLDREKQAAVRQIRKLQEDAELQRQLQMQKQ 180
> bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing
factor 3B subunit 1
Length=1147
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 45/72 (62%), Gaps = 8/72 (11%)
Query 46 ALLERRAIAAREDMYRRQRFQRALSPERQDPFAADGGRRADVSARSYADVMVEQQLDREK 105
LL ++++ ARED YRRQRF R LSPER DPFA + AD R+YAD+M E ++ R +
Sbjct 68 VLLNKKSVLAREDSYRRQRFDRKLSPERYDPFA----KEADPDERTYADIMKETEIMRTR 123
Query 106 QAAVKQIRKLQE 117
K+I E
Sbjct 124 ----KEIEHFME 131
> dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1;
K12828 splicing factor 3B subunit 1
Length=1315
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/55 (52%), Positives = 39/55 (70%), Gaps = 3/55 (5%)
Query 53 IAAREDMYRRQRFQRALSPERQDPFAADGGRRAD--VSARSYADVMVEQQLDREK 105
I+ RED Y+++R + +SPER DPF ADGG+ D V R+Y DVM EQQL +E+
Sbjct 114 ISDREDEYKKRRQKMIISPERHDPF-ADGGKTPDPKVQVRTYMDVMKEQQLSKEE 167
> xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155;
splicing factor 3b, subunit 1, 155kDa; K12828 splicing factor
3B subunit 1
Length=1307
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 44/62 (70%), Gaps = 3/62 (4%)
Query 53 IAAREDMYRRQRFQRALSPERQDPFAADGGRRAD--VSARSYADVMVEQQLDREKQAAVK 110
IA RED Y++QR + +SPER DPF ADGG+ D ++AR++ DVM EQ L +E++ +
Sbjct 110 IANREDEYKQQRRKMIISPERLDPF-ADGGKTPDPKLNARTFKDVMQEQYLTKEEREIRQ 168
Query 111 QI 112
QI
Sbjct 169 QI 170
> mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP155,
SF3b155, TA-8, Targ4; splicing factor 3b, subunit 1; K12828
splicing factor 3B subunit 1
Length=1304
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/62 (48%), Positives = 42/62 (67%), Gaps = 3/62 (4%)
Query 53 IAAREDMYRRQRFQRALSPERQDPFAADGGRRAD--VSARSYADVMVEQQLDREKQAAVK 110
IA RED Y++ R +SPER DPF ADGG+ D ++AR+Y DVM EQ L +E++ +
Sbjct 112 IADREDEYKKHRRTMIISPERLDPF-ADGGKTPDPKMNARTYMDVMREQHLTKEEREIRQ 170
Query 111 QI 112
Q+
Sbjct 171 QL 172
> ath:AT5G64270 splicing factor, putative; K12828 splicing factor
3B subunit 1
Length=1269
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 38/63 (60%), Gaps = 1/63 (1%)
Query 50 RRAIAAREDMYRRQRFQRALSPERQDPFAADGGRRADVSARSYADVMVEQQLDREKQAAV 109
R++IA RE YR +R R LSP+R D FA G + D S R+Y D M E L REK+ +
Sbjct 96 RQSIAEREGEYRNRRLNRVLSPDRVDAFAM-GDKTPDASVRTYTDHMRETALQREKEETM 154
Query 110 KQI 112
+ I
Sbjct 155 RLI 157
> pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828
splicing factor 3B subunit 1
Length=1386
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/92 (22%), Positives = 47/92 (51%), Gaps = 3/92 (3%)
Query 46 ALLERRAIAAREDMYRRQRFQRALSPERQDPFAADGGRRADVSARSYADVMVEQQLDREK 105
L + ++I RE+ ++++++ LSP+R DPFA + R+Y D+M+E + +
Sbjct 111 GLTKDKSIKKRENAFQKKKYDYTLSPQRADPFA---DKSPSPGERTYTDIMLENKKKSKM 167
Query 106 QAAVKQIRKLQEDAAIAQQLLQQRQQEEAAGG 137
+ A +++ + ++ + E + GG
Sbjct 168 KEASRKLSHYNKQGDSTNDTKEKMENENSHGG 199
> tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing
factor 3B subunit 1
Length=1107
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 29/82 (35%), Positives = 47/82 (57%), Gaps = 3/82 (3%)
Query 51 RAIAAREDMYRRQRFQRALSPERQDPFAADGGRRADVSARSYADVMVEQQLDREKQAAVK 110
++I RED YRRQR ++ LSPER DPF+ G+ R++ADVM E ++ R++ K
Sbjct 19 KSIYEREDDYRRQRLRQRLSPERYDPFS---GKTPLPEERTFADVMKETEISRQRNEISK 75
Query 111 QIRKLQEDAAIAQQLLQQRQQE 132
I K + + + + R+ +
Sbjct 76 HISKHGMTKEVEEAIEETRRSK 97
> hsa:23451 SF3B1, Hsh155, PRP10, PRPF10, SAP155, SF3b155; splicing
factor 3b, subunit 1, 155kDa; K12828 splicing factor 3B
subunit 1
Length=144
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 53 IAAREDMYRRQRFQRALSPERQDPFA 78
IA RED Y++ R +SPER DPFA
Sbjct 112 IADREDEYKKHRRTMIISPERLDPFA 137
> cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B
subunit 1
Length=1322
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query 51 RAIAAREDMYRRQRFQRALSPERQDPFAADGGRRADVSARSYADVMVEQQLDREK 105
+ IA R+ Y + +R +SP+R D F + D R YA+VM +Q EK
Sbjct 115 KTIAERQSKYHERAMRRQISPDRADAFV---DQTPDNRNRGYAEVMRDQMYHEEK 166
> cel:C35C5.1 sdc-2; Sex determination and Dosage Compensation
defect family member (sdc-2)
Length=2962
Score = 28.9 bits (63), Expect = 6.0, Method: Composition-based stats.
Identities = 26/105 (24%), Positives = 46/105 (43%), Gaps = 14/105 (13%)
Query 28 SERRAMLASSGGDAGDAAALLERRAIAAREDMYRRQRFQRALSPERQDPFAADGGR---- 83
+E+R M+AS+ LE+R + D QR +R ++ F + R
Sbjct 1087 AEQRRMMAST--------RRLEKRTTQKQVDPETIQRLRREDEVRKRKRFEEEDRRGMIR 1138
Query 84 --RADVSARSYADVMVEQQLDREKQAAVKQIRKLQEDAAIAQQLL 126
V+ + D M+E+ L EK ++IR+ QE+ I + +
Sbjct 1139 RREERVALQEKVDRMLEEGLRLEKVREAERIRQQQEEERIEMETI 1183
Lambda K H
0.314 0.128 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40