bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3140_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
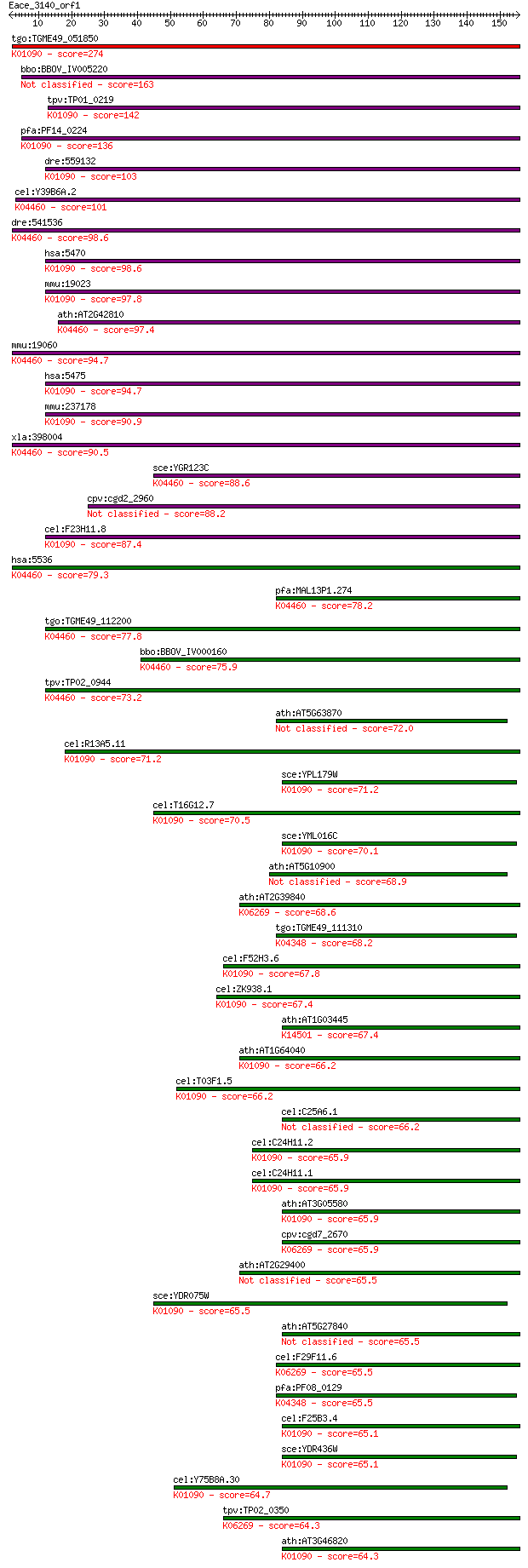
tgo:TGME49_051850 serine/threonine protein phosphatase 5, puta... 274 9e-74
bbo:BBOV_IV005220 23.m06074; serine/threonin protein phosphata... 163 2e-40
tpv:TP01_0219 serine/threonine protein phosphatase; K01090 pro... 142 3e-34
pfa:PF14_0224 PP7; serine/threonine protein phosphatase; K0109... 136 3e-32
dre:559132 ppef1, MGC171580, zgc:171580; protein phosphatase, ... 103 2e-22
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 101 8e-22
dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801 ... 98.6 6e-21
hsa:5470 PPEF2, PPP7CB; protein phosphatase, EF-hand calcium b... 98.6 8e-21
mmu:19023 Ppef2; protein phosphatase, EF hand calcium-binding ... 97.8 1e-20
ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphop... 97.4 2e-20
mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalyt... 94.7 1e-19
hsa:5475 PPEF1, PP7, PPEF, PPP7C, PPP7CA; protein phosphatase,... 94.7 1e-19
mmu:237178 Ppef1, PPEF-1; protein phosphatase with EF hand cal... 90.9 2e-18
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 90.5 2e-18
sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphat... 88.6 8e-18
cpv:cgd2_2960 phosphoprotein phosphatase related 88.2 9e-18
cel:F23H11.8 pef-1; Phosphatase with EF hands family member (p... 87.4 2e-17
hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein ph... 79.3 4e-15
pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase (E... 78.2 1e-14
tgo:TGME49_112200 serine/threonine protein phosphatase, putati... 77.8 1e-14
bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphat... 75.9 6e-14
tpv:TP02_0944 serine/threonine protein phosphatase; K04460 pro... 73.2 3e-13
ath:AT5G63870 PP7; PP7 (SERINE/THREONINE PHOSPHATASE 7); prote... 72.0 7e-13
cel:R13A5.11 hypothetical protein; K01090 protein phosphatase ... 71.2 1e-12
sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein ph... 71.2 1e-12
cel:T16G12.7 hypothetical protein; K01090 protein phosphatase ... 70.5 2e-12
sce:YML016C PPZ1; Ppz1p (EC:3.1.3.16); K01090 protein phosphat... 70.1 3e-12
ath:AT5G10900 calcineurin-like phosphoesterase family protein 68.9 5e-12
ath:AT2G39840 TOPP4; TOPP4; protein serine/threonine phosphata... 68.6 9e-12
tgo:TGME49_111310 serine/threonine protein phosphatase, putati... 68.2 1e-11
cel:F52H3.6 hypothetical protein; K01090 protein phosphatase [... 67.8 1e-11
cel:ZK938.1 hypothetical protein; K01090 protein phosphatase [... 67.4 2e-11
ath:AT1G03445 BSU1; BSU1 (BRI1 SUPPRESSOR 1); protein serine/t... 67.4 2e-11
ath:AT1G64040 TOPP3; TOPP3; protein serine/threonine phosphata... 66.2 3e-11
cel:T03F1.5 gsp-4; yeast Glc Seven-like Phosphatases family me... 66.2 4e-11
cel:C25A6.1 hypothetical protein 66.2 4e-11
cel:C24H11.2 hypothetical protein; K01090 protein phosphatase ... 65.9 5e-11
cel:C24H11.1 hypothetical protein; K01090 protein phosphatase ... 65.9 5e-11
ath:AT3G05580 serine/threonine protein phosphatase, putative; ... 65.9 5e-11
cpv:cgd7_2670 serine/threonine protein phosphatase ; K06269 pr... 65.9 5e-11
ath:AT2G29400 TOPP1; TOPP1 (TYPE ONE PROTEIN PHOSPHATASE 1); p... 65.5 6e-11
sce:YDR075W PPH3; Catalytic subunit of an evolutionarily conse... 65.5 7e-11
ath:AT5G27840 TOPP8; TOPP8; protein serine/threonine phosphatase 65.5 7e-11
cel:F29F11.6 gsp-1; yeast Glc Seven-like Phosphatases family m... 65.5 8e-11
pfa:PF08_0129 serine/threonine protein phosphatase, putative; ... 65.5 8e-11
cel:F25B3.4 hypothetical protein; K01090 protein phosphatase [... 65.1 9e-11
sce:YDR436W PPZ2; Ppz2p (EC:3.1.3.16); K01090 protein phosphat... 65.1 9e-11
cel:Y75B8A.30 pph-4.1; Protein PHosphatase family member (pph-... 64.7 1e-10
tpv:TP02_0350 serine/threonine protein phosphatase; K06269 pro... 64.3 1e-10
ath:AT3G46820 TOPP5; TOPP5; protein serine/threonine phosphata... 64.3 1e-10
> tgo:TGME49_051850 serine/threonine protein phosphatase 5, putative
(EC:3.1.2.15 3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=1086
Score = 274 bits (700), Expect = 9e-74, Method: Compositional matrix adjust.
Identities = 122/154 (79%), Positives = 139/154 (90%), Gaps = 0/154 (0%)
Query 2 IDASTIAEEGIPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHY 61
+DAS+I+ E IP Y+GP + WPITREF LDL+DH+R++PDVPLPRKYALEL+MA+ +HY
Sbjct 246 VDASSISAENIPDDYAGPRVKWPITREFCLDLVDHYRDNPDVPLPRKYALELIMAITDHY 305
Query 62 KQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRG 121
++TMKGAV EVEIPKK G RLVLVGDTHGQLNDVLWI YKFGPPSATNVYLFNGDIADRG
Sbjct 306 RETMKGAVVEVEIPKKGGSRLVLVGDTHGQLNDVLWIFYKFGPPSATNVYLFNGDIADRG 365
Query 122 RSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
R AVEIF++LFAFKL CP SV+INRGNHESADMN
Sbjct 366 RYAVEIFMMLFAFKLQCPSSVVINRGNHESADMN 399
> bbo:BBOV_IV005220 23.m06074; serine/threonin protein phosphatase
(EC:3.1.3.16)
Length=952
Score = 163 bits (413), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 80/156 (51%), Positives = 106/156 (67%), Gaps = 5/156 (3%)
Query 5 STIAEEGIP-----SGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEE 59
S + EE P + Y+GP L +T +FA +L+D ++ +P +YA +++ + E
Sbjct 157 SDVFEESDPQKYPETTYNGPKLGSKVTSKFANELLDAYKSGQISTVPIEYAQRILVDMLE 216
Query 60 HYKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIAD 119
Y+ GA+ VE+P G L++VGD HGQLND+LWI +KFGPPS+ NVYLFNGDIAD
Sbjct 217 WYRSNDNGAINTVEVPPHENGNLIVVGDLHGQLNDLLWIFFKFGPPSSRNVYLFNGDIAD 276
Query 120 RGRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
RG A +IF+LLF+FKL P SVIINRGNHES DMN
Sbjct 277 RGMGATDIFMLLFSFKLADPSSVIINRGNHESEDMN 312
> tpv:TP01_0219 serine/threonine protein phosphatase; K01090 protein
phosphatase [EC:3.1.3.16]
Length=935
Score = 142 bits (359), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 73/143 (51%), Positives = 88/143 (61%), Gaps = 20/143 (13%)
Query 13 PSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAEV 72
PS Y+GP L +T +FA +L+D ++ + +G + +
Sbjct 173 PSDYNGPKLGKVVTLKFAHELLDMYKSGAN--------------------NNDEGVINHI 212
Query 73 EIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLF 132
+IP L++VGD HGQLND+LWI YKFGPPS NVYLFNGDIADRG A IFLLLF
Sbjct 213 DIPTHENSNLIIVGDLHGQLNDLLWIFYKFGPPSRRNVYLFNGDIADRGTFASNIFLLLF 272
Query 133 AFKLNCPGSVIINRGNHESADMN 155
AFKL P SVIINRGNHES DMN
Sbjct 273 AFKLAEPTSVIINRGNHESDDMN 295
> pfa:PF14_0224 PP7; serine/threonine protein phosphatase; K01090
protein phosphatase [EC:3.1.3.16]
Length=959
Score = 136 bits (342), Expect = 3e-32, Method: Composition-based stats.
Identities = 66/152 (43%), Positives = 99/152 (65%), Gaps = 1/152 (0%)
Query 5 STIAEEGIPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQT 64
ST + E + P L I R FA ++ F ++ LP ++++ ++ ++
Sbjct 163 STTSYESSNYNENVPRLKDKIDRTFATEMFHFFLTSKEIILPYNLVHKVLIKTKKMLEEN 222
Query 65 MKGAVAEVEIPKKPGG-RLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRS 123
+K +V +++ KK +L+++GD HGQL+DVLW+ +FG PS+TN+Y+FNGDIADRG +
Sbjct 223 IKSSVINLDMSKKSKDTKLIILGDVHGQLHDVLWLFNRFGIPSSTNIYIFNGDIADRGEN 282
Query 124 AVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
A EIF+LLF FKL+C SVIINRGNHE + MN
Sbjct 283 ATEIFILLFIFKLSCNDSVIINRGNHECSYMN 314
> dre:559132 ppef1, MGC171580, zgc:171580; protein phosphatase,
EF-hand calcium binding domain 1 (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=718
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 57/144 (39%), Positives = 82/144 (56%), Gaps = 6/144 (4%)
Query 12 IPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAE 71
+P YSGP L++P++ L+ F+E + L KY L+L+ ++ KQ
Sbjct 109 VPESYSGPRLSFPLSVTDTNALLSAFKE--EQTLHGKYVLQLLYETKKFLKQMPNVIYLS 166
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
K+ + + GD HG+L+D+L I YK G PSA N Y+FNGD DRG+ + EI ++L
Sbjct 167 TSYAKE----ITICGDLHGRLDDLLLIFYKNGLPSAENPYIFNGDFVDRGKKSTEIIIIL 222
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
FAF L P + +NRGNHE MN
Sbjct 223 FAFVLLYPDHMHLNRGNHEDHIMN 246
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 101 bits (252), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 61/159 (38%), Positives = 78/159 (49%), Gaps = 12/159 (7%)
Query 3 DASTIAEE------GIPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMA 56
D T+AE I Y GP L IT+EF L LI F+ L +KYA ++++
Sbjct 157 DKKTVAETLDINAMAIEDSYDGPRLEDKITKEFVLQLIKTFKNQQ--KLHKKYAFKMLL- 213
Query 57 VEEHYKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 116
+ +K VEI G + + GD HGQ D+ I G PS TN YLFNGD
Sbjct 214 ---EFYNYVKSLPTMVEITVPTGKKFTICGDVHGQFYDLCNIFEINGYPSETNPYLFNGD 270
Query 117 IADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
DRG +VE + FKL P ++RGNHES MN
Sbjct 271 FVDRGSFSVETIFTMIGFKLLYPNHFFMSRGNHESDVMN 309
> dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=481
Score = 98.6 bits (244), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 61/155 (39%), Positives = 85/155 (54%), Gaps = 7/155 (4%)
Query 2 IDASTIAEEGIPSGYSGPHLT-WPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEH 60
+D+ I I Y+GP L +T +F DL+D F++ L RK A ++++ V
Sbjct 144 VDSLDIENMTIEDDYAGPKLEDGKVTLKFMEDLMDWFKDQK--KLHRKCAYQILVQV--- 198
Query 61 YKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADR 120
K+ + + VEI K ++ + GDTHGQ D+L I PS TN YLFNGD DR
Sbjct 199 -KEVLTKLPSLVEITLKETEKITICGDTHGQYYDLLNIFELNSLPSPTNPYLFNGDFVDR 257
Query 121 GRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
G +VE+ L LF FKL P + RGNHE+ +MN
Sbjct 258 GSFSVEVILTLFGFKLLYPEHFHLLRGNHETDNMN 292
> hsa:5470 PPEF2, PPP7CB; protein phosphatase, EF-hand calcium
binding domain 2 (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=753
Score = 98.6 bits (244), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 53/144 (36%), Positives = 79/144 (54%), Gaps = 6/144 (4%)
Query 12 IPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAE 71
+P Y+GP L++P+ + A L++ FR L +Y L L+ ++H Q
Sbjct 110 VPDSYTGPRLSFPLLPDHATALVEAFRLKQQ--LHARYVLNLLYETKKHLVQLPNIN--- 164
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
+ + + GD HGQL+D+++I YK G PS Y+FNGD DRG+ +VEI ++L
Sbjct 165 -RVSTCYSEEITVCGDLHGQLDDLIFIFYKNGLPSPERSYVFNGDFVDRGKDSVEILMIL 223
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
FAF L P +NRGNHE +N
Sbjct 224 FAFMLVYPKEFHLNRGNHEDHMVN 247
> mmu:19023 Ppef2; protein phosphatase, EF hand calcium-binding
domain 2 (EC:3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=757
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 52/144 (36%), Positives = 79/144 (54%), Gaps = 6/144 (4%)
Query 12 IPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAE 71
+P Y+GP L++P+ + A L++ FR L +Y L L+ +H Q
Sbjct 110 VPDSYTGPRLSFPLLPDHATALVEAFRLRQQ--LHARYVLNLLYETRKHLAQLPNINRVS 167
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
++ + + GD HGQL+D+++I YK G PS Y+FNGD DRG+ +VE+ ++L
Sbjct 168 TCYSEE----VTVCGDLHGQLDDLIFIFYKNGLPSPERAYVFNGDFVDRGKDSVEVLMVL 223
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
FAF L P +NRGNHE +N
Sbjct 224 FAFMLVYPKEFHLNRGNHEDHLVN 247
> ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphoprotein
phosphatase/ protein binding / protein serine/threonine
phosphatase; K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=538
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 55/141 (39%), Positives = 77/141 (54%), Gaps = 7/141 (4%)
Query 16 YSGPHLTWP-ITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAEVEI 74
YSG + +T +F +++ F+ L ++YA ++V+ +Q + + V+I
Sbjct 216 YSGARIEGEEVTLDFVKTMMEDFKNQK--TLHKRYAYQIVLQT----RQILLALPSLVDI 269
Query 75 PKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAF 134
G + + GD HGQ D+L I G PS N YLFNGD DRG +VEI L LFAF
Sbjct 270 SVPHGKHITVCGDVHGQFYDLLNIFELNGLPSEENPYLFNGDFVDRGSFSVEIILTLFAF 329
Query 135 KLNCPGSVIINRGNHESADMN 155
K CP S+ + RGNHES MN
Sbjct 330 KCMCPSSIYLARGNHESKSMN 350
> mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalytic
subunit (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=499
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 59/155 (38%), Positives = 82/155 (52%), Gaps = 7/155 (4%)
Query 2 IDASTIAEEGIPSGYSGPHLT-WPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEH 60
+D+ I I YSGP L +T F DL+ +++ L RK A ++++ V
Sbjct 162 VDSLDIESMTIEDEYSGPKLEDGKVTITFMKDLMQWYKDQK--KLHRKCAYQILVQV--- 216
Query 61 YKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADR 120
K+ + VE K ++ + GDTHGQ D+L I G PS TN Y+FNGD DR
Sbjct 217 -KEVLCKLSTLVETTLKETEKITVCGDTHGQFYDLLNIFELNGLPSETNPYIFNGDFVDR 275
Query 121 GRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
G +VE+ L LF FKL P + RGNHE+ +MN
Sbjct 276 GSFSVEVILTLFGFKLLYPDHFHLLRGNHETDNMN 310
> hsa:5475 PPEF1, PP7, PPEF, PPP7C, PPP7CA; protein phosphatase,
EF-hand calcium binding domain 1 (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=653
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 52/144 (36%), Positives = 77/144 (53%), Gaps = 6/144 (4%)
Query 12 IPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAE 71
+P Y+GP L +P+T L++ F+E L Y LE++ K+ +K
Sbjct 103 VPDSYNGPRLQFPLTCTDIDLLLEAFKEQQ--ILHAHYVLEVLFET----KKVLKQMPNF 156
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
I P + + GD HG+L+D+ I YK G PS N Y+FNGD DRG++++EI ++L
Sbjct 157 THIQTSPSKEVTICGDLHGKLDDLFLIFYKNGLPSERNPYVFNGDFVDRGKNSIEILMIL 216
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
L P + +NRGNHE MN
Sbjct 217 CVSFLVYPNDLHLNRGNHEDFMMN 240
> mmu:237178 Ppef1, PPEF-1; protein phosphatase with EF hand calcium-binding
domain 1 (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=650
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 50/144 (34%), Positives = 75/144 (52%), Gaps = 6/144 (4%)
Query 12 IPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAE 71
+P Y GP L +P+T L+ F++ L Y LE++ + KQ +
Sbjct 106 VPDSYDGPRLQFPLTFTDIHILLQAFKQQQ--ILHAHYVLEVLFEARKVLKQMPNFS--- 160
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
+ P + + GD HG+L+D++ I YK G PS N Y+FNGD DRG +++EI ++L
Sbjct 161 -HVKTFPAKEITICGDLHGKLDDLMLIFYKNGLPSENNPYVFNGDFVDRGNNSMEILMIL 219
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
L P + +NRGNHE MN
Sbjct 220 LVCFLVYPSDLHLNRGNHEDFMMN 243
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 53/155 (34%), Positives = 83/155 (53%), Gaps = 7/155 (4%)
Query 2 IDASTIAEEGIPSGYSGPHLT-WPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEH 60
+D+ I I Y+GP L +T +F L+L+ +++ L RK ++++ V
Sbjct 156 VDSLDIEGMTIEDEYTGPQLQDGNVTVDFMLELMQFYKDQK--KLHRKCLYQMLVQV--- 210
Query 61 YKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADR 120
K + + VE+ + ++ + GDTHGQ D++ I + G PS N Y+FNGD DR
Sbjct 211 -KDILSKLPSLVEVSLEKSQQVTVCGDTHGQFYDLMNIFHLNGLPSENNPYIFNGDFVDR 269
Query 121 GRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
G +VE+ + LF FKL P + RGNHE+ MN
Sbjct 270 GSFSVEVIVTLFGFKLLYPAHFHLLRGNHETDTMN 304
> sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphatase
5 [EC:3.1.3.16]
Length=513
Score = 88.6 bits (218), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 43/111 (38%), Positives = 65/111 (58%), Gaps = 2/111 (1%)
Query 45 LPRKYALELVMAVEEHYKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGP 104
LP+KY ++ + ++Q + ++ E+E P ++ + GDTHGQ DVL + KFG
Sbjct 209 LPKKYVAAIISHADTLFRQ--EPSMVELENNSTPDVKISVCGDTHGQFYDVLNLFRKFGK 266
Query 105 PSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
+ YLFNGD DRG + E+ LL + K+ P + +NRGNHES +MN
Sbjct 267 VGPKHTYLFNGDFVDRGSWSCEVALLFYCLKILHPNNFFLNRGNHESDNMN 317
> cpv:cgd2_2960 phosphoprotein phosphatase related
Length=525
Score = 88.2 bits (217), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 51/131 (38%), Positives = 73/131 (55%), Gaps = 3/131 (2%)
Query 25 ITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAEVEIPKKPGGRLVL 84
I F +LID F ++P+ L RKYA +V + + K+ + + I K+ + +
Sbjct 210 IRESFVTELID-FLKNPENRLHRKYAYMIVYDLIQVLKEVASKPLVRINIGKQE--HITV 266
Query 85 VGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVII 144
GD HGQ D+L I G PS N YLFNGD DRG +VE+ L+LF K+ P V +
Sbjct 267 CGDIHGQFFDLLNIFDINGLPSVNNGYLFNGDFVDRGSFSVEVILVLFTLKIMYPYHVHL 326
Query 145 NRGNHESADMN 155
RGNHE+ ++N
Sbjct 327 ARGNHETKNLN 337
> cel:F23H11.8 pef-1; Phosphatase with EF hands family member
(pef-1); K01090 protein phosphatase [EC:3.1.3.16]
Length=707
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 75/144 (52%), Gaps = 6/144 (4%)
Query 12 IPSGYSGPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAVAE 71
I Y GP L+ P+ + +I+ F+ + L KY V+ + ++ K +
Sbjct 187 IDRNYKGPTLSLPLDKPQVAKMIEAFKVNK--VLHPKY----VLMILHEARKIFKAMPSV 240
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
I ++ + GD HG+ +D+ ILYK G PS N Y+FNGD DRG ++E+ +L
Sbjct 241 SRISTSISNQVTICGDLHGKFDDLCIILYKNGYPSVDNPYIFNGDFVDRGGQSIEVLCVL 300
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
FA + P S+ +NRGNHE MN
Sbjct 301 FALVIVDPMSIYLNRGNHEDHIMN 324
> hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein
phosphatase 5, catalytic subunit (EC:3.1.3.16); K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=477
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 56/155 (36%), Positives = 73/155 (47%), Gaps = 29/155 (18%)
Query 2 IDASTIAEEGIPSGYSGPHLT-WPITREFALDLIDHFREHPDVPLPRKYALELVMAVEEH 60
+D+ I I YSGP L +T F +L+ +++ L RK A
Sbjct 162 VDSLDIESMTIEDEYSGPKLEDGKVTISFMKELMQWYKDQK--KLHRKCAY--------- 210
Query 61 YKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADR 120
QT K + + GDTHGQ D+L I G PS TN Y+FNGD DR
Sbjct 211 --QTEK---------------ITVCGDTHGQFYDLLNIFELNGLPSETNPYIFNGDFVDR 253
Query 121 GRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
G +VE+ L LF FKL P + RGNHE+ +MN
Sbjct 254 GSFSVEVILTLFGFKLLYPDHFHLLRGNHETDNMN 288
> pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=658
Score = 78.2 bits (191), Expect = 1e-14, Method: Composition-based stats.
Identities = 36/74 (48%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 82 LVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGS 141
L + GD HGQ D+L I+ G PS N YLFNGD DRG +VE+ + L+ KL P +
Sbjct 398 LTICGDVHGQYYDLLNIMKINGYPSEKNSYLFNGDFVDRGSFSVEVIIFLYLAKLTFPNN 457
Query 142 VIINRGNHESADMN 155
V + RGNHE+ +MN
Sbjct 458 VYLTRGNHETDNMN 471
> tgo:TGME49_112200 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 50/149 (33%), Positives = 75/149 (50%), Gaps = 11/149 (7%)
Query 12 IPSGYSGPHLTWPITREFA----LDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKG 67
+ Y GP + P E+ + + F + L +++A +V+ V ++ G
Sbjct 219 VEKSYDGP-VYAPTGSEYCSAEFVGALKEFLKDTSKRLHKQFAYRMVLDV-----ISLLG 272
Query 68 AVAEVEIPKKPG-GRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVE 126
+ +E + P G + + GD HGQ D+ I G PS N YLFNGD DRG +VE
Sbjct 273 KLQTLERIRIPDDGEITVCGDVHGQYYDLCNIFELNGMPSKENPYLFNGDFVDRGSFSVE 332
Query 127 IFLLLFAFKLNCPGSVIINRGNHESADMN 155
LLL+A KL P + + RGNHE+ +MN
Sbjct 333 CILLLYAAKLAFPDAFHLTRGNHETFEMN 361
> bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphatase
5 (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=545
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 44/115 (38%), Positives = 62/115 (53%), Gaps = 4/115 (3%)
Query 41 PDVPLPRKYALELVMAVEEHYKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILY 100
P L RKY +++++ V K+ +V ++ + +K + + GD HGQ D+L I
Sbjct 247 PGNTLHRKYVIQIILDVINILKEY--DSVVDLSLTEKDD--ITICGDIHGQFYDLLNIFE 302
Query 101 KFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
G PS TN YLFNGD DRG +VE + LF K P I RGNHE+ +N
Sbjct 303 INGSPSETNGYLFNGDFVDRGSFSVECAIALFLAKAFYPKKFFITRGNHETEALN 357
> tpv:TP02_0944 serine/threonine protein phosphatase; K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 49/146 (33%), Positives = 67/146 (45%), Gaps = 9/146 (6%)
Query 12 IPSGYSGPHLTW--PITREFALDLIDHFREHPDVPLPRKYALELVMAVEEHYKQTMKGAV 69
+ GY+GP ++ P+ L + E+ V L + +HY Q + V
Sbjct 222 VDQGYTGPVFSFKEPMNHLDKSYLFHNTLEYIKVTL----TFLFYLGPWKHYSQEYESVV 277
Query 70 AEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFL 129
++ P L + GD HGQ D+L I G PS N YLFNGD DRG + E
Sbjct 278 ---DVNVYPSDELTVCGDIHGQFYDLLNIFSINGEPSDENSYLFNGDFVDRGSFSFECVF 334
Query 130 LLFAFKLNCPGSVIINRGNHESADMN 155
LF K+ P S I RGNHE+ +N
Sbjct 335 TLFLAKVLFPSSFHIVRGNHETEALN 360
> ath:AT5G63870 PP7; PP7 (SERINE/THREONINE PHOSPHATASE 7); protein
serine/threonine phosphatase
Length=301
Score = 72.0 bits (175), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 34/70 (48%), Positives = 47/70 (67%), Gaps = 0/70 (0%)
Query 82 LVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGS 141
+V+VGD HGQL+D+L++L G P Y+FNGD DRG +E FL+L ++K+ P
Sbjct 79 VVVVGDIHGQLHDLLFLLKDTGFPCQNRCYVFNGDYVDRGAWGLETFLVLLSWKVLMPDR 138
Query 142 VIINRGNHES 151
V + RGNHES
Sbjct 139 VYLLRGNHES 148
> cel:R13A5.11 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=421
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/144 (31%), Positives = 74/144 (51%), Gaps = 13/144 (9%)
Query 18 GPHLTWPITREFALDLIDHFREHPDVPLPRKYALELVMAVEE---HYKQTM---KGAVAE 71
GP +T + +D +D H V + ++ E + QT+ + A+ +
Sbjct 91 GPEMTAQNRTQERIDFVDFLDRHYQVIQAGVHKIQYTKEEFENVIYEAQTIFSSEKALVD 150
Query 72 VEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLL 131
++ P V+VGD HGQ ND++ + G P T VY+F GD DRG ++E +LL
Sbjct 151 IDPP------CVVVGDLHGQFNDLINMFILLGRPPET-VYVFTGDYVDRGMMSLECIMLL 203
Query 132 FAFKLNCPGSVIINRGNHESADMN 155
F +K+ P ++++ RGNHE A +N
Sbjct 204 FTYKICYPENIVLLRGNHEIARVN 227
> sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=549
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 48/71 (67%), Gaps = 1/71 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+VGD HGQ ND+L IL G PS TN YLF GD DRG++++E LLL +K+ +
Sbjct 298 VVGDVHGQFNDLLRILKLSGVPSDTN-YLFLGDYVDRGKNSLETILLLLCYKIKYKDNFF 356
Query 144 INRGNHESADM 154
+ RGNHESA++
Sbjct 357 MLRGNHESANV 367
> cel:T16G12.7 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=319
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/111 (36%), Positives = 64/111 (57%), Gaps = 9/111 (8%)
Query 45 LPRKYALELVMAVEEHYKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGP 104
+ +K + L+ A ++ + + +GA+ E+E P K + GD HGQ +DVL + + G
Sbjct 40 MSQKTLVSLLDAAKDVFMK--QGAMLELEAPVK------ICGDVHGQYSDVLRLFDRGGF 91
Query 105 PSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
P N YLF GD DRG+ ++E+ L A+K+ PG+ + RGNHE +N
Sbjct 92 PPLVN-YLFLGDYVDRGQQSLEVACLFLAYKVKFPGNFFMLRGNHECGSIN 141
> sce:YML016C PPZ1; Ppz1p (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=692
Score = 70.1 bits (170), Expect = 3e-12, Method: Composition-based stats.
Identities = 33/71 (46%), Positives = 47/71 (66%), Gaps = 1/71 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+VGD HGQ D+L + K G P ++N YLF GD DRG+ ++E LLLF +K+ P +
Sbjct 416 IVGDVHGQYGDLLRLFTKCGFPPSSN-YLFLGDYVDRGKQSLETILLLFCYKIKYPENFF 474
Query 144 INRGNHESADM 154
+ RGNHE A++
Sbjct 475 LLRGNHECANV 485
> ath:AT5G10900 calcineurin-like phosphoesterase family protein
Length=600
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 50/72 (69%), Gaps = 0/72 (0%)
Query 80 GRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCP 139
R+++VGD HGQL+D+L I + G PS ++FNG+ RG ++E+FL+L A+K+ P
Sbjct 223 SRVIVVGDLHGQLHDLLKIFDQSGRPSQNQCFVFNGNYIGRGSWSLEVFLVLLAWKIMMP 282
Query 140 GSVIINRGNHES 151
+VI+ RG+ E+
Sbjct 283 ENVILLRGSSET 294
> ath:AT2G39840 TOPP4; TOPP4; protein serine/threonine phosphatase;
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=321
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 54/87 (62%), Gaps = 11/87 (12%)
Query 71 EVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGDIADRGRSAVEIF 128
E+E P K + GD HGQ +D+L L+++G PPSA YLF GD DRG+ ++E
Sbjct 64 ELEAPIK------ICGDIHGQYSDLLR-LFEYGGFPPSAN--YLFLGDYVDRGKQSLETI 114
Query 129 LLLFAFKLNCPGSVIINRGNHESADMN 155
LL A+K+ PG+ + RGNHE A +N
Sbjct 115 CLLLAYKIKYPGNFFLLRGNHECASIN 141
> tgo:TGME49_111310 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic
subunit [EC:3.1.3.16]
Length=501
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 45/73 (61%), Gaps = 1/73 (1%)
Query 82 LVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGS 141
+ +VGD HGQ D+L +L G P T YLF GD DRG ++E+ LLL+A KLN P
Sbjct 82 ITVVGDIHGQFYDLLKLLDVGGDPDTTQ-YLFLGDYVDRGSFSIEVLLLLYAIKLNHPTR 140
Query 142 VIINRGNHESADM 154
V + RGNHE M
Sbjct 141 VWLLRGNHECRQM 153
> cel:F52H3.6 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=329
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/90 (40%), Positives = 48/90 (53%), Gaps = 7/90 (7%)
Query 66 KGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAV 125
+ + EV+ P + + GD HGQ D+L I + P N YLF GD DRGR +
Sbjct 45 QATMVEVQAP------IAVCGDIHGQYTDLLRIFNRCSFPPDQN-YLFLGDYVDRGRQQL 97
Query 126 EIFLLLFAFKLNCPGSVIINRGNHESADMN 155
E+ LL A+K+ P I RGNHE A +N
Sbjct 98 EVICLLMAYKIRYPNRFFILRGNHECASIN 127
> cel:ZK938.1 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=327
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 49/92 (53%), Gaps = 7/92 (7%)
Query 64 TMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRS 123
+ + + EV+ P + + GD HGQ D+L I + P N YLF GD DRGR
Sbjct 43 SAQATMVEVQAP------IAVCGDIHGQYTDLLRIFNRCSFPPDQN-YLFLGDYVDRGRQ 95
Query 124 AVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
+E+ LL A+K+ P I RGNHE A +N
Sbjct 96 QLEVICLLMAYKVRYPNGFFILRGNHECASIN 127
> ath:AT1G03445 BSU1; BSU1 (BRI1 SUPPRESSOR 1); protein serine/threonine
phosphatase; K14501 serine/threonine-protein phosphatase
BSU1 [EC:3.1.3.16]
Length=793
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 51/77 (66%), Gaps = 5/77 (6%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSA----TNV-YLFNGDIADRGRSAVEIFLLLFAFKLNC 138
+ GD HGQ D++ + +++G PS T++ YLF GD DRG+ ++EI +LLFA K+
Sbjct 507 VFGDIHGQYGDLMRLFHEYGHPSVEGDITHIDYLFLGDYVDRGQHSLEIIMLLFALKIEY 566
Query 139 PGSVIINRGNHESADMN 155
P ++ + RGNHES MN
Sbjct 567 PKNIHLIRGNHESLAMN 583
> ath:AT1G64040 TOPP3; TOPP3; protein serine/threonine phosphatase;
K01090 protein phosphatase [EC:3.1.3.16]
Length=322
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 49/85 (57%), Gaps = 7/85 (8%)
Query 71 EVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLL 130
E+E P K + GDTHGQ +D+L + G P A N YLF GD DRG+ +VE L
Sbjct 51 ELEAPIK------ICGDTHGQFSDLLRLFEYGGYPPAAN-YLFLGDYVDRGKQSVETICL 103
Query 131 LFAFKLNCPGSVIINRGNHESADMN 155
L A+K+ + + RGNHE A +N
Sbjct 104 LLAYKIKYKENFFLLRGNHECASIN 128
> cel:T03F1.5 gsp-4; yeast Glc Seven-like Phosphatases family
member (gsp-4); K01090 protein phosphatase [EC:3.1.3.16]
Length=305
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 57/114 (50%), Gaps = 17/114 (14%)
Query 52 ELVMAVEEHYKQTM----------KGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYK 101
L +V E QT + ++ EVE P +++ GD HGQ +D+L I K
Sbjct 23 RLTTSVNEQELQTCCAVAKSVFASQASLLEVEPP------IIVCGDIHGQYSDLLRIFDK 76
Query 102 FGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHESADMN 155
G P N +LF GD DRGR +E L+F FK+ P + + RGNHE +N
Sbjct 77 NGFPPDIN-FLFLGDYVDRGRQNIETICLMFCFKIKYPENFFMLRGNHECPAIN 129
> cel:C25A6.1 hypothetical protein
Length=300
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 46/72 (63%), Gaps = 1/72 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+ GD HGQ D+L + ++ G P N YLF GD DRGR ++E +LL A+K+ PG++
Sbjct 58 VCGDIHGQFPDLLRLFHRGGWPPTAN-YLFLGDYVDRGRFSIETIVLLLAYKVKFPGNLF 116
Query 144 INRGNHESADMN 155
+ RGNHE +N
Sbjct 117 LLRGNHECEFVN 128
> cel:C24H11.2 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=384
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 48/89 (53%), Gaps = 9/89 (10%)
Query 75 PKKPGGRLVL--------VGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVE 126
P KP +L GDTHGQ ND+L I G + T YLF GD DRG ++E
Sbjct 99 PPKPAANALLELQAPINICGDTHGQYNDLLRIFNACGAATKTQ-YLFLGDYVDRGGHSLE 157
Query 127 IFLLLFAFKLNCPGSVIINRGNHESADMN 155
+ +LLF+ KL P + + RGNHE +N
Sbjct 158 VIMLLFSLKLAMPRKMHLLRGNHELKAIN 186
> cel:C24H11.1 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=384
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 48/89 (53%), Gaps = 9/89 (10%)
Query 75 PKKPGGRLVL--------VGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVE 126
P KP +L GDTHGQ ND+L I G + T YLF GD DRG ++E
Sbjct 99 PPKPAANALLELQAPINICGDTHGQYNDLLRIFNACGAATKTQ-YLFLGDYVDRGGHSLE 157
Query 127 IFLLLFAFKLNCPGSVIINRGNHESADMN 155
+ +LLF+ KL P + + RGNHE +N
Sbjct 158 VIMLLFSLKLAMPRKMHLLRGNHELKAIN 186
> ath:AT3G05580 serine/threonine protein phosphatase, putative;
K01090 protein phosphatase [EC:3.1.3.16]
Length=318
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 47/74 (63%), Gaps = 5/74 (6%)
Query 84 LVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGS 141
+ GD HGQ D+L L+++G PPSA YLF GD DRG+ ++E LL A+K+ P
Sbjct 63 ICGDIHGQYQDLLR-LFEYGGYPPSAN--YLFLGDYVDRGKQSLETICLLLAYKIRYPSK 119
Query 142 VIINRGNHESADMN 155
+ + RGNHE A +N
Sbjct 120 IFLLRGNHEDAKIN 133
> cpv:cgd7_2670 serine/threonine protein phosphatase ; K06269
protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=320
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+ GD HGQ D+L + G P TN YLF GD DRG+ ++E LLFA+K+ P +
Sbjct 75 ICGDIHGQYYDLLRLFEYGGFPPDTN-YLFLGDYVDRGKQSLETICLLFAYKIKYPENFF 133
Query 144 INRGNHESADMN 155
+ RGNHE A +N
Sbjct 134 LLRGNHECASIN 145
> ath:AT2G29400 TOPP1; TOPP1 (TYPE ONE PROTEIN PHOSPHATASE 1);
protein serine/threonine phosphatase
Length=318
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/87 (43%), Positives = 53/87 (60%), Gaps = 11/87 (12%)
Query 71 EVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGDIADRGRSAVEIF 128
E+E P K + GD HGQ +D+L L+++G PP A YLF GD DRG+ ++E
Sbjct 67 ELEAPIK------ICGDIHGQYSDLLR-LFEYGGFPPEAN--YLFLGDYVDRGKQSLETI 117
Query 129 LLLFAFKLNCPGSVIINRGNHESADMN 155
LL A+K+ P + + RGNHESA +N
Sbjct 118 CLLLAYKIKYPENFFLLRGNHESASIN 144
> sce:YDR075W PPH3; Catalytic subunit of an evolutionarily conserved
protein phosphatase complex containing Psy2p and the
regulatory subunit Psy4p; required for cisplatin resistance;
involved in activation of Gln3p (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=308
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 57/107 (53%), Gaps = 9/107 (8%)
Query 45 LPRKYALELVMAVEEHYKQTMKGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGP 104
+P + L + +E +G V +V+ P + + GD HGQL+D+L + K G
Sbjct 17 IPEETVFRLCLNSQELLMN--EGNVTQVDTP------VTICGDIHGQLHDLLTLFEKSGG 68
Query 105 PSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHES 151
T Y+F GD DRG ++E FLLL +KL P + + RGNHE+
Sbjct 69 VEKTR-YIFLGDFVDRGFYSLESFLLLLCYKLRYPDRITLIRGNHET 114
> ath:AT5G27840 TOPP8; TOPP8; protein serine/threonine phosphatase
Length=324
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 47/74 (63%), Gaps = 5/74 (6%)
Query 84 LVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGS 141
+ GD HGQ D+L L+++G PPSA YLF GD DRG+ ++E LL A+K+ P
Sbjct 63 ICGDIHGQYQDLLR-LFEYGGYPPSAN--YLFLGDYVDRGKQSLETICLLLAYKIRYPSK 119
Query 142 VIINRGNHESADMN 155
+ + RGNHE A +N
Sbjct 120 IYLLRGNHEDAKIN 133
> cel:F29F11.6 gsp-1; yeast Glc Seven-like Phosphatases family
member (gsp-1); K06269 protein phosphatase 1, catalytic subunit
[EC:3.1.3.16]
Length=329
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 35/76 (46%), Positives = 48/76 (63%), Gaps = 5/76 (6%)
Query 82 LVLVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCP 139
L + GD HGQ ND+L L+++G PP A YLF GD DRG+ ++E LL A+K+ P
Sbjct 59 LKICGDIHGQYNDLLR-LFEYGGFPPEAN--YLFLGDYVDRGKQSLETICLLLAYKVKYP 115
Query 140 GSVIINRGNHESADMN 155
+ + RGNHE A +N
Sbjct 116 ENFFLLRGNHECASIN 131
> pfa:PF08_0129 serine/threonine protein phosphatase, putative;
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 45/73 (61%), Gaps = 1/73 (1%)
Query 82 LVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGS 141
+ +VGD HGQ D+L +L G P T +LF GD DRG ++E+ LLL+A K+N P
Sbjct 82 ITIVGDIHGQYYDLLKLLEVGGNPDHTQ-FLFLGDYVDRGSFSIEVLLLLYALKINFPDR 140
Query 142 VIINRGNHESADM 154
+ + RGNHE M
Sbjct 141 IWLIRGNHECRQM 153
> cel:F25B3.4 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=363
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+ GD HGQ +D+L + K G P N YLF GD DRG+ +E LLLFA+K+ P
Sbjct 110 ICGDIHGQFSDLLRLFDKNGFPHRAN-YLFLGDYVDRGKHCLETILLLFAYKVIFPNHFF 168
Query 144 INRGNHESADMN 155
+ RGNHE + +N
Sbjct 169 MLRGNHECSLIN 180
> sce:YDR436W PPZ2; Ppz2p (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=710
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 44/71 (61%), Gaps = 1/71 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+VGD HGQ D+L + K G P N YLF GD DRG+ ++E LLL +K+ P +
Sbjct 451 IVGDVHGQYADLLRLFTKCGFPPMAN-YLFLGDYVDRGKQSLETILLLLCYKIKYPENFF 509
Query 144 INRGNHESADM 154
+ RGNHE A++
Sbjct 510 LLRGNHECANV 520
> cel:Y75B8A.30 pph-4.1; Protein PHosphatase family member (pph-4.1);
K01090 protein phosphatase [EC:3.1.3.16]
Length=333
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/108 (39%), Positives = 57/108 (52%), Gaps = 8/108 (7%)
Query 51 LELVMAVEEHYKQTMKGAVAEV-EIPKKPGGRLVL------VGDTHGQLNDVLWILYKFG 103
+E +M E +Q +K A+ EI + G V+ GD HGQ D++ L+K G
Sbjct 36 IEKLMRCELIAEQDVKTLCAKAREILAEEGNVQVIDSPVTICGDIHGQFYDLM-ELFKVG 94
Query 104 PPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVIINRGNHES 151
P YLF GD DRG +VE FLLL A K P +++ RGNHES
Sbjct 95 GPVPNTNYLFLGDFVDRGFYSVETFLLLLALKARYPDRMMLIRGNHES 142
> tpv:TP02_0350 serine/threonine protein phosphatase; K06269 protein
phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=301
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/90 (40%), Positives = 50/90 (55%), Gaps = 7/90 (7%)
Query 66 KGAVAEVEIPKKPGGRLVLVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAV 125
+ + E+E P K + GD HGQ D+L + G P A N YLF GD DRG+ ++
Sbjct 45 QSILLELEAPIK------ICGDIHGQYYDLLRLFEYGGFPPAAN-YLFLGDYVDRGKQSL 97
Query 126 EIFLLLFAFKLNCPGSVIINRGNHESADMN 155
E LL A+K+ P + + RGNHE A +N
Sbjct 98 ETICLLLAYKIKYPENFFLLRGNHECASIN 127
> ath:AT3G46820 TOPP5; TOPP5; protein serine/threonine phosphatase;
K01090 protein phosphatase [EC:3.1.3.16]
Length=312
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 84 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGDIADRGRSAVEIFLLLFAFKLNCPGSVI 143
+ GD HGQ +D+L + G P A N YLF GD DRG+ ++E LL A+K+ P +
Sbjct 67 ICGDIHGQYSDLLRLFEYGGFPPAAN-YLFLGDYVDRGKQSLETICLLLAYKIKYPENFF 125
Query 144 INRGNHESADMN 155
+ RGNHE A +N
Sbjct 126 LLRGNHECASIN 137
Lambda K H
0.320 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40