bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3267_orf3
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
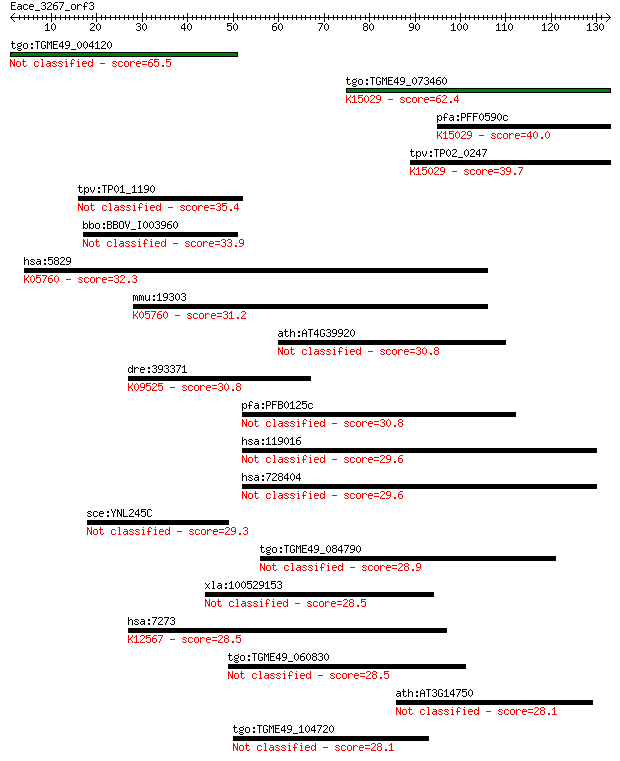
tgo:TGME49_004120 hypothetical protein 65.5 5e-11
tgo:TGME49_073460 eukaryotic translation initiation factor 3 s... 62.4 4e-10
pfa:PFF0590c homologue of human HSPC025; K15029 translation in... 40.0 0.002
tpv:TP02_0247 hypothetical protein; K15029 translation initiat... 39.7 0.002
tpv:TP01_1190 hypothetical protein 35.4 0.040
bbo:BBOV_I003960 19.m02294; hypothetical protein 33.9 0.15
hsa:5829 PXN, FLJ16691; paxillin; K05760 paxillin 32.3 0.35
mmu:19303 Pxn, AW108311, AW123232, Pax; paxillin; K05760 paxillin 31.2 0.88
ath:AT4G39920 POR; POR (PORCINO); binding 30.8 1.1
dre:393371 dnajc5gb, MGC63689, wu:fa01g12, zgc:63689; DnaJ (Hs... 30.8 1.2
pfa:PFB0125c conserved Plasmodium protein 30.8 1.3
hsa:119016 AGAP4, CTGLF1, MRIP2; ArfGAP with GTPase domain, an... 29.6 2.2
hsa:728404 AGAP8, CTGLF5; ArfGAP with GTPase domain, ankyrin r... 29.6 2.2
sce:YNL245C CWC25; Cwc25p 29.3 3.2
tgo:TGME49_084790 hypothetical protein 28.9 4.1
xla:100529153 rfx6; regulatory factor X, 6 28.5
hsa:7273 TTN, CMD1G, CMH9, CMPD4, DKFZp451N061, EOMFC, FLJ2602... 28.5 5.0
tgo:TGME49_060830 hypothetical protein 28.5 5.2
ath:AT3G14750 hypothetical protein 28.1 6.7
tgo:TGME49_104720 SWIM zinc finger domain-containing protein (... 28.1 7.2
> tgo:TGME49_004120 hypothetical protein
Length=475
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 32/50 (64%), Positives = 40/50 (80%), Gaps = 0/50 (0%)
Query 1 LGQLQAQAGLLTKTQAQVAAAGLEWLYEDPKNSAEAKEQEREAFLLGKPI 50
L +LQA AGL++K A VAAAGLEWLY++PK EA+E REA+LLGKP+
Sbjct 54 LRKLQADAGLISKQAAVVAAAGLEWLYDEPKGQKEAQEAAREAYLLGKPL 103
> tgo:TGME49_073460 eukaryotic translation initiation factor 3
subunit 6 interacting protein, putative ; K15029 translation
initiation factor 3 subunit L
Length=639
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/59 (55%), Positives = 40/59 (67%), Gaps = 1/59 (1%)
Query 75 AAAEQQQRERVEAE-LADEDLDRMMAERVAFEPEVENFLRELYDKLYIRDADAIRRLYE 132
+ Q + VE E L D LDRMM E+V FE +VE FLR LYD++Y R+ DAIRRLYE
Sbjct 40 VSPSNQASQNVEGEGLDDAALDRMMGEKVRFENDVEEFLRSLYDEVYDRNVDAIRRLYE 98
> pfa:PFF0590c homologue of human HSPC025; K15029 translation
initiation factor 3 subunit L
Length=656
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 95 DRMMAERVAFEPEVENFLRELYDKLYIRDADAIRRLYE 132
+ ++ E V+FE EVE FL L+D +Y R+A+AI++L++
Sbjct 9 ENVIEEIVSFEEEVETFLINLHDFVYHRNAEAIKKLFD 46
> tpv:TP02_0247 hypothetical protein; K15029 translation initiation
factor 3 subunit L
Length=544
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 29/44 (65%), Gaps = 0/44 (0%)
Query 89 LADEDLDRMMAERVAFEPEVENFLRELYDKLYIRDADAIRRLYE 132
LA + R + + + EPEV +FL L+D LY ++A+A++ LYE
Sbjct 11 LATDGPQRTVTQLLPIEPEVVDFLVRLHDNLYRKNAEALKNLYE 54
> tpv:TP01_1190 hypothetical protein
Length=346
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 4/36 (11%)
Query 16 AQVAAAGLEWLYEDPKNSAEAKEQEREAFLLGKPIP 51
+++ A GLEWLY DP ++ E++LLG+ IP
Sbjct 10 SKIEAEGLEWLYNDP----TQQQNNLESYLLGESIP 41
> bbo:BBOV_I003960 19.m02294; hypothetical protein
Length=511
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query 17 QVAAAGLEWLYEDPKNSAEAKEQEREAFLLGKPI 50
+V A GLEWLY DP ++ ++ + E +LLGK I
Sbjct 22 RVEAEGLEWLYADP-SANKSNANQLEEYLLGKSI 54
> hsa:5829 PXN, FLJ16691; paxillin; K05760 paxillin
Length=591
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 29/103 (28%), Positives = 45/103 (43%), Gaps = 16/103 (15%)
Query 4 LQAQAGLLTKTQAQV-AAAGLEWLYEDPKNSAEAKEQEREAFLLGKPIPNVAPAADESIE 62
L +AG LTK + + GLE D + S E+ E E+ +P+ PA
Sbjct 190 LGGKAGPLTKEKPKRNGGRGLE----DVRPSVESLLDELES-----SVPSPVPAIT---- 236
Query 63 KRVGESSLKAPAAAAEQQQRERVEAELADEDLDRMMAERVAFE 105
V + + +P QQ+ R+ A A +LD +MA F+
Sbjct 237 --VNQGEMSSPQRVTSTQQQTRISASSATRELDELMASLSDFK 277
> mmu:19303 Pxn, AW108311, AW123232, Pax; paxillin; K05760 paxillin
Length=557
Score = 31.2 bits (69), Expect = 0.88, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 11/78 (14%)
Query 28 EDPKNSAEAKEQEREAFLLGKPIPNVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEA 87
ED + S E+ E E+ +P+ PA V + + +P QQ+ R+ A
Sbjct 211 EDVRPSVESLLDELES-----SVPSPVPAIT------VNQGEMSSPQRVTSSQQQTRISA 259
Query 88 ELADEDLDRMMAERVAFE 105
A +LD +MA F+
Sbjct 260 SSATRELDELMASLSDFK 277
> ath:AT4G39920 POR; POR (PORCINO); binding
Length=345
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 60 SIEKRVGESSLKAPAAAAEQQQRERVEAELADEDLDRMMAERVAFEPEVE 109
SIE R+ ES L + + + + + + E +A ++L++++AE F P E
Sbjct 66 SIESRIAESRLASSSTDSSKLKSDLAEISVAIDNLEKLLAENSYFLPSYE 115
> dre:393371 dnajc5gb, MGC63689, wu:fa01g12, zgc:63689; DnaJ (Hsp40)
homolog, subfamily C, member 5 gamma b; K09525 DnaJ homolog
subfamily C member 5
Length=211
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 27 YEDPKNSAEAKEQER--EAFLLGKPIPNVAPAADESIEKRVG 66
Y DP+ AE +E+ R E ++G+PIP+ APAAD VG
Sbjct 145 YVDPEELAEEEERSRDNETIIIGQPIPSPAPAADSGYPVIVG 186
> pfa:PFB0125c conserved Plasmodium protein
Length=1341
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Query 52 NVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEAELADEDLDRMMAERVAFEPEVENF 111
N +E +++VG++ + +++ RE + AE+ +E D M E + FE ENF
Sbjct 573 NFFEEVEEKYDEKVGKNIFEEVEEKFDEKMRENIFAEIEEEKYDEKMGENI-FEEVEENF 631
> hsa:119016 AGAP4, CTGLF1, MRIP2; ArfGAP with GTPase domain,
ankyrin repeat and PH domain 4
Length=663
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 33/84 (39%), Gaps = 9/84 (10%)
Query 52 NVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEAELADEDL------DRMMAERVAFE 105
+V P+ E E G+ AP AAA Q VE EDL DR M E + F
Sbjct 23 SVCPSESEIYEAGAGDRMAGAPMAAAVQPAEVTVE---VGEDLHMHHVRDREMPEALEFN 79
Query 106 PEVENFLRELYDKLYIRDADAIRR 129
P ++ + D IRR
Sbjct 80 PSANPEASTIFQRNSQTDVVEIRR 103
> hsa:728404 AGAP8, CTGLF5; ArfGAP with GTPase domain, ankyrin
repeat and PH domain 8
Length=663
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 33/84 (39%), Gaps = 9/84 (10%)
Query 52 NVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEAELADEDL------DRMMAERVAFE 105
+V P+ E E G+ AP AAA Q VE EDL DR M E + F
Sbjct 23 SVCPSESEIYEAGAGDRMAGAPMAAAVQPAEVTVE---VGEDLHMHHVRDREMPEALEFN 79
Query 106 PEVENFLRELYDKLYIRDADAIRR 129
P ++ + D IRR
Sbjct 80 PSANPEASTIFQRNSQTDVVEIRR 103
> sce:YNL245C CWC25; Cwc25p
Length=179
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 5/31 (16%)
Query 18 VAAAGLEWLYEDPKNSAEAKEQEREAFLLGK 48
+ +GLEW+Y+D K S E+E +LLGK
Sbjct 71 LKKSGLEWMYQDAKLS-----DEKEDYLLGK 96
> tgo:TGME49_084790 hypothetical protein
Length=976
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 30/65 (46%), Gaps = 2/65 (3%)
Query 56 AADESIEKRVGESSLKAPAAAAEQQQRERVEAELADEDLDRMMAERVAFEPEVENFLREL 115
A E + + GE + PA + QRE V+ +LA L + A+ + E+ E
Sbjct 590 AKAEELYQTCGELIMMQPATINREGQREIVDEKLA--HLKELEAQLASLHQEISTLANEA 647
Query 116 YDKLY 120
DK++
Sbjct 648 TDKVF 652
> xla:100529153 rfx6; regulatory factor X, 6
Length=926
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 44 FLLGKPIPNVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEAELADED 93
F G+P+P VA D+ VG+ L P +Q RE + +ED
Sbjct 16 FQNGEPLPKVASGMDDCYGTMVGKDLLSYPQEDLCEQGREGIMLTPEEED 65
> hsa:7273 TTN, CMD1G, CMH9, CMPD4, DKFZp451N061, EOMFC, FLJ26020,
FLJ26409, FLJ32040, FLJ34413, FLJ39564, FLJ43066, HMERF,
LGMD2J, TMD; titin (EC:2.7.11.1); K12567 titin [EC:2.7.11.1]
Length=26926
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 32/70 (45%), Gaps = 4/70 (5%)
Query 27 YEDPKNSAEAKEQEREAFLLGKPIPNVAPAADESIEKRVGESSLKAPAAAAEQQQRERVE 86
Y P+ + +E ++ IP A A+ E R A AA + +QR+R+
Sbjct 25645 YSSPQAHVKVEETRKDFRYSTYHIPTKAEASTSYAELR----ERHAQAAYRQPKQRQRIM 25700
Query 87 AELADEDLDR 96
AE DE+L R
Sbjct 25701 AEREDEELLR 25710
> tgo:TGME49_060830 hypothetical protein
Length=1081
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 49 PIPNVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEAELADEDLDRMMAE 100
P +V P A E I R GE PA +E+ +R+ DE+ M+A+
Sbjct 356 PTLSVLPEAAERINSRDGEEPAGHPAGVSEEGSVQRLLCGHTDEENPEMLAD 407
> ath:AT3G14750 hypothetical protein
Length=331
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 5/48 (10%)
Query 86 EAELADEDLDRMMAERVAFEPEVENFLRELYDK-----LYIRDADAIR 128
E E+A +L R+M + E E +RE+YDK + +R+ DA+R
Sbjct 101 ELEVAQHELQRIMHYIDSLRAEEEIMMREMYDKSMRSEMELREVDAMR 148
> tgo:TGME49_104720 SWIM zinc finger domain-containing protein
(EC:2.1.1.43)
Length=6036
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 50 IPNVAPAADESIEKRVGESSLKAPAAAAEQQQRERVEAELADE 92
IP PAAD S + R G + APA + E + V ++ +DE
Sbjct 4908 IPTCVPAADSS-QARGGNGGVSAPAFSKETAEETAVTSDESDE 4949
Lambda K H
0.311 0.128 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40