bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3308_orf2
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
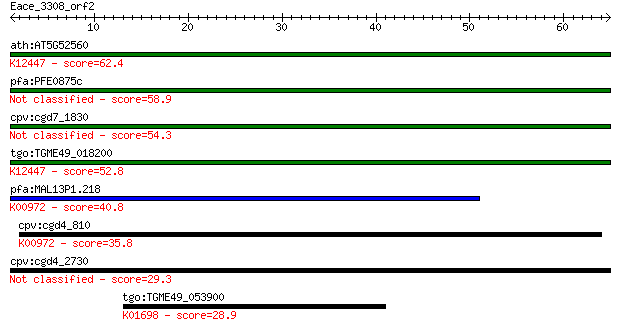
ath:AT5G52560 ATUSP; ATUSP (ARABIDOPSIS THALIANA UDP-SUGAR PYR... 62.4 3e-10
pfa:PFE0875c conserved Plasmodium protein, unknown function 58.9 4e-09
cpv:cgd7_1830 secreted UDP-N-acetylglucosamine pyrophosphoryla... 54.3 1e-07
tgo:TGME49_018200 UDP-N-acetylglucosamine pyrophosphorylase, p... 52.8 3e-07
pfa:MAL13P1.218 UDP-N-acetylglucosamine pyrophosphorylase, put... 40.8 0.001
cpv:cgd4_810 UDP-N-acetylglucosamine pyrophosphorylase ; K0097... 35.8 0.030
cpv:cgd4_2730 coiled-coil protein 29.3 3.3
tgo:TGME49_053900 delta-aminolevulinic acid dehydratase, putat... 28.9 4.4
> ath:AT5G52560 ATUSP; ATUSP (ARABIDOPSIS THALIANA UDP-SUGAR PYROPHOSPHORYLASE);
UTP-monosaccharide-1-phosphate uridylyltransferase/
UTP:arabinose-1-phosphate uridylyltransferase/ UTP:galactose-1-phosphate
uridylyltransferase/ UTP:glucose-1-phosphate
uridylyltransferase/ UTP:xylose-1-phosphate uridylyltransferase/
glucuronate-1-phosphate uridylyltransferase; K12447
UDP-sugar pyrophosphorylase [EC:2.7.7.64]
Length=614
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 32/66 (48%), Positives = 41/66 (62%), Gaps = 2/66 (3%)
Query 1 FVAGGLGERLGYNGIKLSLPVESCTETSYIEYYCSFIRAFQEYAAAKTKKAVH--IPLAL 58
VAGGLGERLGYNGIK++LP E+ T T ++++Y I A QE + IP +
Sbjct 133 LVAGGLGERLGYNGIKVALPRETTTGTCFLQHYIESILALQEASNKIDSDGSERDIPFII 192
Query 59 MTSADT 64
MTS DT
Sbjct 193 MTSDDT 198
> pfa:PFE0875c conserved Plasmodium protein, unknown function
Length=855
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 30/64 (46%), Positives = 43/64 (67%), Gaps = 0/64 (0%)
Query 1 FVAGGLGERLGYNGIKLSLPVESCTETSYIEYYCSFIRAFQEYAAAKTKKAVHIPLALMT 60
+AGGLGERL + IK+ L +E +YIEYYC++IR F++Y + KK ++IP +M
Sbjct 288 LLAGGLGERLKHKDIKIKLFTNLISEETYIEYYCNYIRCFEKYIKKEKKKKMNIPFIIML 347
Query 61 SADT 64
S DT
Sbjct 348 SDDT 351
> cpv:cgd7_1830 secreted UDP-N-acetylglucosamine pyrophosphorylase
family protein, signal peptide
Length=654
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 38/64 (59%), Gaps = 0/64 (0%)
Query 1 FVAGGLGERLGYNGIKLSLPVESCTETSYIEYYCSFIRAFQEYAAAKTKKAVHIPLALMT 60
VAGGLGERL + GIK+ + + + ++ + Y ++IR +Q + + IPL +MT
Sbjct 148 LVAGGLGERLAFEGIKIGIELSMASNITFFQLYTNYIREYQRRLKEAFGEDIVIPLLIMT 207
Query 61 SADT 64
S DT
Sbjct 208 SDDT 211
> tgo:TGME49_018200 UDP-N-acetylglucosamine pyrophosphorylase,
putative ; K12447 UDP-sugar pyrophosphorylase [EC:2.7.7.64]
Length=655
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 32/92 (34%), Positives = 44/92 (47%), Gaps = 28/92 (30%)
Query 1 FVAGGLGERLGYNGIKLSLPVESCTETSYIEYYCSFIRAFQ-----------EYAAAKTK 49
VAGGLGERLGY GIK+ LP E+ T ++ + YC ++ + Q E A ++
Sbjct 109 LVAGGLGERLGYKGIKIGLPCETSTGKTFAQLYCEYLLSIQSGLAREAASGEEEVEATSE 168
Query 50 KAVH-----------------IPLALMTSADT 64
A +PLA+MTS DT
Sbjct 169 GASEEGRGEAVNSVSAGRSPCVPLAIMTSDDT 200
> pfa:MAL13P1.218 UDP-N-acetylglucosamine pyrophosphorylase, putative
(EC:2.7.7.23); K00972 UDP-N-acetylglucosamine pyrophosphorylase
[EC:2.7.7.23]
Length=593
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 1 FVAGGLGERLGYNGIKLSLPVESCTETSYIEYYCSFIRAFQEYAAAKTKK 50
F+AGGLG RLG N K+ L V T ++++++ I QEY + KK
Sbjct 111 FLAGGLGSRLGLNKPKVLLEVTPLTNKTFLQFFFEQIIFLQEYCSLYEKK 160
> cpv:cgd4_810 UDP-N-acetylglucosamine pyrophosphorylase ; K00972
UDP-N-acetylglucosamine pyrophosphorylase [EC:2.7.7.23]
Length=603
Score = 35.8 bits (81), Expect = 0.030, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 36/78 (46%), Gaps = 16/78 (20%)
Query 2 VAGGLGERLGYNGIKLSLPVESCTETSYIEYYCSFIRAFQEYAAAKT------------- 48
++GG G RLGYNG K P+ ++ S+ + +C I++ + +
Sbjct 193 MSGGDGSRLGYNGPKGMYPIGKISKDSFFKIFCQKIQSLIRLVSKENYDHDTDDLKSKET 252
Query 49 ---KKAVHIPLALMTSAD 63
K+ IPL +MTS +
Sbjct 253 KYLKEMKEIPLYIMTSEN 270
> cpv:cgd4_2730 coiled-coil protein
Length=987
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 34/75 (45%), Gaps = 13/75 (17%)
Query 1 FVAGGLGERLGYNGIKLSLPVESCTETSY-----------IEYYCSFIRAFQEYAAAKTK 49
FV GL E +G+N +L P ++ SY +EYY + I + +Y
Sbjct 125 FVPNGL-EIVGFNSTELDQPKDNLISNSYLEEKNLGFSGQLEYYVNNIESI-DYGQKSGI 182
Query 50 KAVHIPLALMTSADT 64
+ + +P+ M + DT
Sbjct 183 QPISVPILRMNTIDT 197
> tgo:TGME49_053900 delta-aminolevulinic acid dehydratase, putative
(EC:4.2.1.24); K01698 porphobilinogen synthase [EC:4.2.1.24]
Length=658
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 13 NGIKLSLPVESCTETSYIEYYCSFIRAF 40
+ I+ SL +E CT+TS + Y C + +F
Sbjct 493 SAIRESLDMEGCTDTSILAYSCKYASSF 520
Lambda K H
0.320 0.134 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2051595972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40