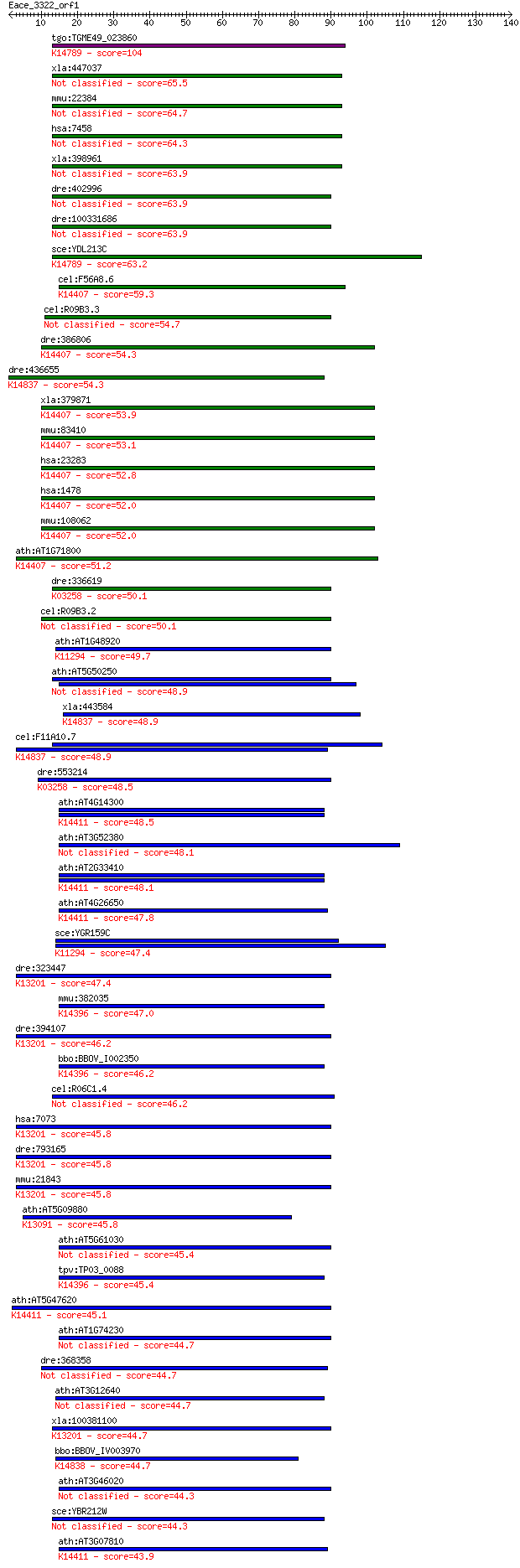
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3322_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_023860 RNA-binding protein, putative ; K14789 nucle... 104 7e-23
xla:447037 eif4h, MGC82967, wbscr1; eukaryotic translation ini... 65.5 5e-11
mmu:22384 Eif4h, AU018978, D5Ertd355e, E430026L18Rik, Ef4h, Wb... 64.7 9e-11
hsa:7458 EIF4H, KIAA0038, WBSCR1, WSCR1; eukaryotic translatio... 64.3 1e-10
xla:398961 hypothetical protein MGC68480 63.9 1e-10
dre:402996 eif4h, MGC77282, wbscr1, zgc:77282; eukaryotic tran... 63.9 1e-10
dre:100331686 eukaryotic translation initiation factor 4H-like 63.9 1e-10
sce:YDL213C NOP6; Nop6p; K14789 nucleolar protein 6 63.2
cel:F56A8.6 cpf-2; Cleavage and Polyadenylation Factor family ... 59.3 3e-09
cel:R09B3.3 hypothetical protein 54.7 9e-08
dre:386806 cstf2, CstF-64, fb11e07, wu:fb11e07, zgc:56346, zgc... 54.3 1e-07
dre:436655 rbm34, zgc:92062; RNA binding motif protein 34; K14... 54.3 1e-07
xla:379871 cstf2, MGC53575, MGC83019, cstf-64; cleavage stimul... 53.9 2e-07
mmu:83410 Cstf2t, 64kDa, C77975, tCstF-64, tauCstF-64; cleavag... 53.1 3e-07
hsa:23283 CSTF2T, CstF-64T, DKFZp434C1013, KIAA0689; cleavage ... 52.8 3e-07
hsa:1478 CSTF2, CstF-64; cleavage stimulation factor, 3' pre-R... 52.0 5e-07
mmu:108062 Cstf2, 64kDa, C630034J23Rik, Cstf64; cleavage stimu... 52.0 6e-07
ath:AT1G71800 cleavage stimulation factor, putative; K14407 cl... 51.2 1e-06
dre:336619 eif4bb, MGC101532, eif4b, fb15c09, wu:fb15c09, zgc:... 50.1 2e-06
cel:R09B3.2 hypothetical protein 50.1 2e-06
ath:AT1G48920 ATNUC-L1; nucleic acid binding / nucleotide bind... 49.7 3e-06
ath:AT5G50250 31 kDa ribonucleoprotein, chloroplast, putative ... 48.9 4e-06
xla:443584 rbm34, MGC115058; RNA binding motif protein 34; K14... 48.9 5e-06
cel:F11A10.7 hypothetical protein; K14837 nucleolar protein 12 48.9
dre:553214 eif4ba, eif4b; eukaryotic translation initiation fa... 48.5 6e-06
ath:AT4G14300 heterogeneous nuclear ribonucleoprotein, putativ... 48.5 7e-06
ath:AT3G52380 CP33; CP33; RNA binding 48.1 7e-06
ath:AT2G33410 heterogeneous nuclear ribonucleoprotein, putativ... 48.1 8e-06
ath:AT4G26650 RNA recognition motif (RRM)-containing protein; ... 47.8 1e-05
sce:YGR159C NSR1, SHE5; Nsr1p; K11294 nucleolin 47.4 1e-05
dre:323447 tia1l, MGC55893, TIA1, fb98f09, wu:fb98f09, zgc:558... 47.4 2e-05
mmu:382035 Pabpn1l, Gm1108, Pabpnl1, ePABP2; poly(A)binding pr... 47.0 2e-05
dre:394107 tial1, MGC66130, zgc:66130; TIA1 cytotoxic granule-... 46.2 3e-05
bbo:BBOV_I002350 19.m02195; RNA binding protein; K14396 polyad... 46.2 3e-05
cel:R06C1.4 hypothetical protein 46.2 3e-05
hsa:7073 TIAL1, MGC33401, TCBP, TIAR; TIA1 cytotoxic granule-a... 45.8 4e-05
dre:793165 tia1, MGC55520, fb73d10, wu:fb73d10, wu:fc02b04, zg... 45.8 4e-05
mmu:21843 Tial1, 5330433G13Rik, AL033329, TIAR, mTIAR; Tia1 cy... 45.8 4e-05
ath:AT5G09880 RNA recognition motif (RRM)-containing protein; ... 45.8 4e-05
ath:AT5G61030 GR-RBP3 (glycine-rich RNA-binding protein 3); AT... 45.4 5e-05
tpv:TP03_0088 hypothetical protein; K14396 polyadenylate-bindi... 45.4 5e-05
ath:AT5G47620 heterogeneous nuclear ribonucleoprotein, putativ... 45.1 7e-05
ath:AT1G74230 GR-RBP5 (glycine-rich RNA-binding protein 5); AT... 44.7 8e-05
dre:368358 sb:cb657 44.7 9e-05
ath:AT3G12640 RNA binding / nucleic acid binding / nucleotide ... 44.7 1e-04
xla:100381100 tial1, tcbp, tiar; TIA1 cytotoxic granule-associ... 44.7 1e-04
bbo:BBOV_IV003970 23.m05815; RNA recognition motif containing ... 44.7 1e-04
ath:AT3G46020 RNA-binding protein, putative 44.3 1e-04
sce:YBR212W NGR1, RBP1; Ngr1p 44.3 1e-04
ath:AT3G07810 heterogeneous nuclear ribonucleoprotein, putativ... 43.9 1e-04
> tgo:TGME49_023860 RNA-binding protein, putative ; K14789 nucleolar
protein 6
Length=247
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 49/81 (60%), Positives = 60/81 (74%), Gaps = 0/81 (0%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
F+LF GNLPL A D+++FF KLQ I +VR+LT ++T KPKGC FVEF KEAL+IA
Sbjct 163 FILFAGNLPLDTKAEDLKAFFKNKLQPRIVEVRMLTHRETNKPKGCAFVEFDCKEALEIA 222
Query 73 LNYDGRTLNERKIRVELTAGG 93
LNY R L RKI +EL+AGG
Sbjct 223 LNYHHRELGGRKINIELSAGG 243
> xla:447037 eif4h, MGC82967, wbscr1; eukaryotic translation initiation
factor 4H
Length=250
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/80 (42%), Positives = 47/80 (58%), Gaps = 3/80 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
F +VGNLP D+ + F L ++ VRL+ DK+T K KG +VEF + E+LK A
Sbjct 42 FTAYVGNLPFNTVQGDIDNIF---KDLSVRSVRLVRDKETDKFKGFCYVEFDDLESLKEA 98
Query 73 LNYDGRTLNERKIRVELTAG 92
L YDG +R IRV++ G
Sbjct 99 LTYDGAIFIDRAIRVDIAEG 118
> mmu:22384 Eif4h, AU018978, D5Ertd355e, E430026L18Rik, Ef4h,
Wbscr1, Wscr1, mKIAA0038; eukaryotic translation initiation
factor 4H
Length=248
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 46/80 (57%), Gaps = 3/80 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
+ +VGNLP D+ + F L I+ VRL+ DK T K KG +VEF ++LK A
Sbjct 42 YTAYVGNLPFNTVQGDIDAIF---KDLSIRSVRLVRDKDTDKFKGFCYVEFDEVDSLKEA 98
Query 73 LNYDGRTLNERKIRVELTAG 92
L YDG L +R +RV++ G
Sbjct 99 LTYDGALLGDRSLRVDIAEG 118
> hsa:7458 EIF4H, KIAA0038, WBSCR1, WSCR1; eukaryotic translation
initiation factor 4H
Length=248
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 46/80 (57%), Gaps = 3/80 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
+ +VGNLP D+ + F L I+ VRL+ DK T K KG +VEF ++LK A
Sbjct 42 YTAYVGNLPFNTVQGDIDAIF---KDLSIRSVRLVRDKDTDKFKGFCYVEFDEVDSLKEA 98
Query 73 LNYDGRTLNERKIRVELTAG 92
L YDG L +R +RV++ G
Sbjct 99 LTYDGALLGDRSLRVDIAEG 118
> xla:398961 hypothetical protein MGC68480
Length=250
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 47/80 (58%), Gaps = 3/80 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
F +VGNLP D+ + F L ++ VRL+ DK+T K KG +VEF + E+LK A
Sbjct 42 FTAYVGNLPFNTVQGDIDNIF---KDLSVRSVRLVRDKETDKFKGFCYVEFDDLESLKEA 98
Query 73 LNYDGRTLNERKIRVELTAG 92
L +DG +R IRV++ G
Sbjct 99 LTFDGAIFIDRAIRVDIAEG 118
> dre:402996 eif4h, MGC77282, wbscr1, zgc:77282; eukaryotic translation
initiation factor 4h
Length=262
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 47/77 (61%), Gaps = 3/77 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
+ +VGNLP D+ + F L ++ VRL+ DK+T K KG +VEF + E+LK A
Sbjct 44 YTAYVGNLPFNTVQGDIDAIF---RDLSVRSVRLVRDKETDKFKGFCYVEFDDLESLKEA 100
Query 73 LNYDGRTLNERKIRVEL 89
L YDG L +R +RV++
Sbjct 101 LTYDGALLGDRSLRVDI 117
> dre:100331686 eukaryotic translation initiation factor 4H-like
Length=262
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 47/77 (61%), Gaps = 3/77 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
+ +VGNLP D+ + F L ++ VRL+ DK+T K KG +VEF + E+LK A
Sbjct 44 YTAYVGNLPFNTVQGDIDAIF---RDLSVRSVRLVRDKETDKFKGFCYVEFDDLESLKEA 100
Query 73 LNYDGRTLNERKIRVEL 89
L YDG L +R +RV++
Sbjct 101 LTYDGALLGDRSLRVDI 117
> sce:YDL213C NOP6; Nop6p; K14789 nucleolar protein 6
Length=225
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 43/113 (38%), Positives = 61/113 (53%), Gaps = 20/113 (17%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEA---- 68
F++FVG+LP ++A ++++ F +RL DK G F+EF +
Sbjct 78 FIVFVGSLPRDITAVELQNHFKNS---SPDQIRLRADK------GIAFLEFDADKDRTGI 128
Query 69 ---LKIALNYDGRTLNERKIRVELTAGGGGKSKIRAEKIKKKN----EERRKR 114
+ IAL G L E+KI VELT GGGG S+ R EK+K KN EER++R
Sbjct 129 QRRMDIALLQHGTLLKEKKINVELTVGGGGNSQERLEKLKNKNIKLDEERKER 181
> cel:F56A8.6 cpf-2; Cleavage and Polyadenylation Factor family
member (cpf-2); K14407 cleavage stimulation factor subunit
2
Length=336
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 50/80 (62%), Gaps = 3/80 (3%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIAL- 73
+FVGN+ VS +RS F + + ++++ D++T KPKG GF+EF + + ++A+
Sbjct 20 VFVGNISYDVSEDTIRSIFSKAGNVL--SIKMVHDRETGKPKGYGFIEFPDIQTAEVAIR 77
Query 74 NYDGRTLNERKIRVELTAGG 93
N +G L+ R +RV+ AGG
Sbjct 78 NLNGYELSGRILRVDSAAGG 97
> cel:R09B3.3 hypothetical protein
Length=85
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 49/80 (61%), Gaps = 3/80 (3%)
Query 11 SLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALK 70
S F ++VGN P + D+ ++F + + +VR++ D++T +P+G FVEFT + A +
Sbjct 2 SGFSVYVGNAPFQTTEDDLGNYFSQAGN--VSNVRIVCDRETGRPRGFAFVEFTEEAAAQ 59
Query 71 IALN-YDGRTLNERKIRVEL 89
A++ ++G N R +RV L
Sbjct 60 RAVDQFNGVDFNGRALRVNL 79
> dre:386806 cstf2, CstF-64, fb11e07, wu:fb11e07, zgc:56346, zgc:77730;
cleavage stimulation factor, 3' pre-RNA, subunit 2;
K14407 cleavage stimulation factor subunit 2
Length=488
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/93 (33%), Positives = 54/93 (58%), Gaps = 3/93 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKE-A 68
+SL +FVGN+P + ++ F ++ L + RL+ D++T KPKG GF E+ ++E A
Sbjct 21 RSLRSVFVGNIPYEATEEQLKDIFS-EVGLVVS-FRLVYDRETGKPKGYGFCEYQDQETA 78
Query 69 LKIALNYDGRTLNERKIRVELTAGGGGKSKIRA 101
L N +GR + R +RV+ A K ++++
Sbjct 79 LSAMRNLNGREFSGRALRVDNAASEKNKEELKS 111
> dre:436655 rbm34, zgc:92062; RNA binding motif protein 34; K14837
nucleolar protein 12
Length=411
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 52/87 (59%), Gaps = 5/87 (5%)
Query 1 AQSSEAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGF 60
+Q S+ K+S+FV GNLP +S +++ F I+ VRL+ D+ + KG G+
Sbjct 243 SQHSKHDHKRSIFV---GNLPYDISELPLQNHFQECGN--IEAVRLVRDRDSGMGKGFGY 297
Query 61 VEFTNKEALKIALNYDGRTLNERKIRV 87
V F + +++ +AL +G TL +RKIRV
Sbjct 298 VLFESPDSVMLALKLNGSTLQQRKIRV 324
> xla:379871 cstf2, MGC53575, MGC83019, cstf-64; cleavage stimulation
factor, 3' pre-RNA, subunit 2, 64kDa; K14407 cleavage
stimulation factor subunit 2
Length=518
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKE-A 68
+SL +FVGN+P + ++ F + RL+ D++T KPKG GF E+ ++E A
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGP--VVSFRLVYDRETGKPKGYGFCEYQDQETA 70
Query 69 LKIALNYDGRTLNERKIRVELTAGGGGKSKIRA 101
L N +GR + R +RV+ A K ++++
Sbjct 71 LSAMRNLNGREFSGRALRVDNAASEKNKEELKS 103
> mmu:83410 Cstf2t, 64kDa, C77975, tCstF-64, tauCstF-64; cleavage
stimulation factor, 3' pre-RNA subunit 2, tau; K14407 cleavage
stimulation factor subunit 2
Length=632
Score = 53.1 bits (126), Expect = 3e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKE-A 68
+SL +FVGN+P + ++ F + RL+ D++T KPKG GF E+ ++E A
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGS--VVSFRLVYDRETGKPKGYGFCEYQDQETA 70
Query 69 LKIALNYDGRTLNERKIRVELTAGGGGKSKIRA 101
L N +GR + R +RV+ A K ++++
Sbjct 71 LSAMRNLNGREFSGRALRVDNAASEKNKEELKS 103
> hsa:23283 CSTF2T, CstF-64T, DKFZp434C1013, KIAA0689; cleavage
stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant;
K14407 cleavage stimulation factor subunit 2
Length=616
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKE-A 68
+SL +FVGN+P + ++ F + RL+ D++T KPKG GF E+ ++E A
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGS--VVSFRLVYDRETGKPKGYGFCEYQDQETA 70
Query 69 LKIALNYDGRTLNERKIRVELTAGGGGKSKIRA 101
L N +GR + R +RV+ A K ++++
Sbjct 71 LSAMRNLNGREFSGRALRVDNAASEKNKEELKS 103
> hsa:1478 CSTF2, CstF-64; cleavage stimulation factor, 3' pre-RNA,
subunit 2, 64kDa; K14407 cleavage stimulation factor subunit
2
Length=577
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKE-A 68
+SL +FVGN+P + ++ F + RL+ D++T KPKG GF E+ ++E A
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGP--VVSFRLVYDRETGKPKGYGFCEYQDQETA 70
Query 69 LKIALNYDGRTLNERKIRVELTAGGGGKSKIRA 101
L N +GR + R +RV+ A K ++++
Sbjct 71 LSAMRNLNGREFSGRALRVDNAASEKNKEELKS 103
> mmu:108062 Cstf2, 64kDa, C630034J23Rik, Cstf64; cleavage stimulation
factor, 3' pre-RNA subunit 2; K14407 cleavage stimulation
factor subunit 2
Length=580
Score = 52.0 bits (123), Expect = 6e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKE-A 68
+SL +FVGN+P + ++ F + RL+ D++T KPKG GF E+ ++E A
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGP--VVSFRLVYDRETGKPKGYGFCEYQDQETA 70
Query 69 LKIALNYDGRTLNERKIRVELTAGGGGKSKIRA 101
L N +GR + R +RV+ A K ++++
Sbjct 71 LSAMRNLNGREFSGRALRVDNAASEKNKEELKS 103
> ath:AT1G71800 cleavage stimulation factor, putative; K14407
cleavage stimulation factor subunit 2
Length=461
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 32/101 (31%), Positives = 52/101 (51%), Gaps = 6/101 (5%)
Query 3 SSEAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVE 62
+S + Q++ +FVGN+P + +R G + RL+TD++T KPKG GF E
Sbjct 2 ASSSSQRR---CVFVGNIPYDATEEQLREICGEVGP--VVSFRLVTDRETGKPKGYGFCE 56
Query 63 FTNKE-ALKIALNYDGRTLNERKIRVELTAGGGGKSKIRAE 102
+ ++E AL N +N R++RV+ G K R +
Sbjct 57 YKDEETALSARRNLQSYEINGRQLRVDFAENDKGTDKTRDQ 97
> dre:336619 eif4bb, MGC101532, eif4b, fb15c09, wu:fb15c09, zgc:101532;
eukaryotic translation initiation factor 4Bb; K03258
translation initiation factor 4B
Length=569
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 43/80 (53%), Gaps = 8/80 (10%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKP---KGCGFVEFTNKEAL 69
+ F+GNLP VS +R FF L I VRL ++ P KG G+ EF + E+L
Sbjct 96 YTAFLGNLPYDVSEESIRDFF---RGLAISAVRLP--REPNNPERLKGFGYAEFDDIESL 150
Query 70 KIALNYDGRTLNERKIRVEL 89
AL+ + L R+IRV++
Sbjct 151 LRALSLNEENLGNRRIRVDI 170
> cel:R09B3.2 hypothetical protein
Length=83
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 46/81 (56%), Gaps = 3/81 (3%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEAL 69
+F ++VGN P + ++ +FF Q I +VR++ D++T +P+G F+EF + +
Sbjct 2 SGIFSVYVGNAPFQTTEEELGNFFSSIGQ--INNVRIVCDRETGRPRGFAFIEFAEEGSA 59
Query 70 KIAL-NYDGRTLNERKIRVEL 89
+ A+ +G N R +RV L
Sbjct 60 QRAVEQMNGAEFNGRPLRVNL 80
> ath:AT1G48920 ATNUC-L1; nucleic acid binding / nucleotide binding;
K11294 nucleolin
Length=557
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Query 14 VLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIAL 73
LF NL + ADV +FF ++ DVR T++ +G G VEF + E + AL
Sbjct 298 TLFAANLSFNIERADVENFFKEAGEVV--DVRFSTNRDDGSFRGFGHVEFASSEEAQKAL 355
Query 74 NYDGRTLNERKIRVEL 89
+ GR L R+IR+++
Sbjct 356 EFHGRPLLGREIRLDI 371
> ath:AT5G50250 31 kDa ribonucleoprotein, chloroplast, putative
/ RNA-binding protein RNP-T, putative / RNA-binding protein
1/2/3, putative / RNA-binding protein cp31, putative
Length=289
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 46/78 (58%), Gaps = 3/78 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
F ++VGNLP V + + F + + D R+++D++T + +G GFV+ +N+ + +A
Sbjct 207 FRIYVGNLPWDVDSGRLERLFSEHGK--VVDARVVSDRETGRSRGFGFVQMSNENEVNVA 264
Query 73 L-NYDGRTLNERKIRVEL 89
+ DG+ L R I+V +
Sbjct 265 IAALDGQNLEGRAIKVNV 282
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEF-TNKEALKIAL 73
LFVGNLP V + + F + + I +V + ++ T + +G GFV T +EA K
Sbjct 115 LFVGNLPYDVDSQALAMLFEQAGTVEISEV--IYNRDTDQSRGFGFVTMSTVEEAEKAVE 172
Query 74 NYDGRTLNERKIRVELTAGGGGK 96
++ +N R++ V A G +
Sbjct 173 KFNSFEVNGRRLTVNRAAPRGSR 195
> xla:443584 rbm34, MGC115058; RNA binding motif protein 34; K14837
nucleolar protein 12
Length=414
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 48/84 (57%), Gaps = 4/84 (4%)
Query 16 FVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALNY 75
FVGNLP + +R F + ++ VR++ D++T KG G+V F + +A+++AL
Sbjct 276 FVGNLPYDIEEESIRKHFSQCGD--VQGVRIIRDQKTGIGKGFGYVLFESADAVQLALKL 333
Query 76 DGRTLNERKIRV--ELTAGGGGKS 97
+ L+ R+IRV +TA KS
Sbjct 334 NNSQLSGRRIRVKRSVTAEAAQKS 357
> cel:F11A10.7 hypothetical protein; K14837 nucleolar protein
12
Length=394
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 49/91 (53%), Gaps = 1/91 (1%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
+FVGNLP ++ + +FF ++ ++ VR++ DK T K KG FV F ++ +A
Sbjct 246 MAIFVGNLPFEITEDALITFFSAQIGP-VEAVRIVRDKDTGKGKGFAFVNFKQDSSVSLA 304
Query 73 LNYDGRTLNERKIRVELTAGGGGKSKIRAEK 103
L+ + + +R +R+ G +KI+ K
Sbjct 305 LSMETIKMEKRDLRITKVMKKGHLTKIQTAK 335
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 49/101 (48%), Gaps = 15/101 (14%)
Query 3 SSEAPQKKSLFVLFVGNLPLTVSAADVRSFFG----------RKL----QLFIKDVRLLT 48
++ A ++ +FVGN+PLT++ VR F R L + K V LT
Sbjct 133 NARASAAENALTVFVGNMPLTMNEKSVRRIFSDFGTISSVRMRNLLPANEKLTKRVTHLT 192
Query 49 DKQTKKPKGCGF-VEFTNKEALKIALNYDGRTLNERKIRVE 88
K K F V+F +E+++ AL Y+G L++ IRV+
Sbjct 193 GKLNDKQSSLTFYVKFGAEESVEKALKYNGTKLDDHVIRVD 233
> dre:553214 eif4ba, eif4b; eukaryotic translation initiation
factor 4Ba; K03258 translation initiation factor 4B
Length=616
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 45/84 (53%), Gaps = 8/84 (9%)
Query 9 KKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKP---KGCGFVEFTN 65
+ + F+GNLP V+ +++FF L I VRL ++ P KG G+ EF +
Sbjct 92 RSPPYTAFLGNLPYDVTEDSIKNFF---RGLSISAVRLP--REPSNPERLKGFGYAEFDD 146
Query 66 KEALKIALNYDGRTLNERKIRVEL 89
E+L AL+ + L R+IRV++
Sbjct 147 VESLLQALSLNEENLGNRRIRVDI 170
> ath:AT4G14300 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=411
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 44/73 (60%), Gaps = 2/73 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
+FVG LP T++ + R +F ++ + DV ++ D+ T +P+G GFV F +++A+ L+
Sbjct 112 IFVGGLPPTLTDEEFRQYF--EVYGPVTDVAIMYDQATNRPRGFGFVSFDSEDAVDSVLH 169
Query 75 YDGRTLNERKIRV 87
L+ +++ V
Sbjct 170 KTFHDLSGKQVEV 182
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
LFVG + +R F + + ++ DK T +P+G GFV F++ L L
Sbjct 8 LFVGGISWETDEDKLREHFTNYGE--VSQAIVMRDKLTGRPRGFGFVIFSDPSVLDRVLQ 65
Query 75 YDGRTLNERKIRV 87
+ +++ R++ V
Sbjct 66 -EKHSIDTREVDV 77
> ath:AT3G52380 CP33; CP33; RNA binding
Length=329
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 53/96 (55%), Gaps = 4/96 (4%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
L+VGNLP T++++++ FG + DV+++ DK T + +G GFV + E K A+
Sbjct 118 LYVGNLPYTITSSELSQIFGEAGT--VVDVQIVYDKVTDRSRGFGFVTMGSIEEAKEAMQ 175
Query 75 -YDGRTLNERKIRVELT-AGGGGKSKIRAEKIKKKN 108
++ + R ++V GG++++ KI+ N
Sbjct 176 MFNSSQIGGRTVKVNFPEVPRGGENEVMRTKIRDNN 211
> ath:AT2G33410 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=404
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
+FVG LP +++ + R++F + + D ++ D+ T++P+G GFV F +++++ + L+
Sbjct 112 IFVGGLPPALTSDEFRAYF--ETYGPVSDAVIMIDQTTQRPRGFGFVSFDSEDSVDLVLH 169
Query 75 YDGRTLNERKIRV 87
LN +++ V
Sbjct 170 KTFHDLNGKQVEV 182
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 36/76 (47%), Gaps = 9/76 (11%)
Query 15 LFVGNLPLTVSAADVRSFF---GRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKI 71
LF+G + +R +F G LQ V ++ +K T +P+G GFV F++ +
Sbjct 8 LFIGGISWDTDENLLREYFSNFGEVLQ-----VTVMREKATGRPRGFGFVAFSDPAVIDR 62
Query 72 ALNYDGRTLNERKIRV 87
L D ++ R + V
Sbjct 63 VLQ-DKHHIDNRDVDV 77
> ath:AT4G26650 RNA recognition motif (RRM)-containing protein;
K14411 RNA-binding protein Musashi
Length=452
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 46/74 (62%), Gaps = 2/74 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
+FVG LP +++ A+ +++F + I DV ++ D T++P+G GF+ F ++E++ + L+
Sbjct 121 IFVGGLPSSITEAEFKNYFDQFGT--IADVVVMYDHNTQRPRGFGFITFDSEESVDMVLH 178
Query 75 YDGRTLNERKIRVE 88
LN + + V+
Sbjct 179 KTFHELNGKMVEVK 192
> sce:YGR159C NSR1, SHE5; Nsr1p; K11294 nucleolin
Length=414
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 44/79 (55%), Gaps = 3/79 (3%)
Query 14 VLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIAL 73
LF+GNL + F + ++ VR+ T +T++PKG G+V+F+N E K AL
Sbjct 268 TLFLGNLSFNADRDAIFELFAKHGEVV--SVRIPTHPETEQPKGFGYVQFSNMEDAKKAL 325
Query 74 N-YDGRTLNERKIRVELTA 91
+ G ++ R +R++ ++
Sbjct 326 DALQGEYIDNRPVRLDFSS 344
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/92 (21%), Positives = 44/92 (47%), Gaps = 3/92 (3%)
Query 14 VLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIAL 73
+FVG L ++ ++ F + R++ ++ T + +G G+V+F NK + A+
Sbjct 169 TIFVGRLSWSIDDEWLKKEFEHIGGVI--GARVIYERGTDRSRGYGYVDFENKSYAEKAI 226
Query 74 N-YDGRTLNERKIRVELTAGGGGKSKIRAEKI 104
G+ ++ R I +++ + RA+K
Sbjct 227 QEMQGKEIDGRPINCDMSTSKPAGNNDRAKKF 258
> dre:323447 tia1l, MGC55893, TIA1, fb98f09, wu:fb98f09, zgc:55893,
zgc:76917; cytotoxic granule-associated RNA binding protein
1, like; K13201 nucleolysin TIA-1/TIAR
Length=342
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 49/91 (53%), Gaps = 6/91 (6%)
Query 3 SSEAPQKK---SLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCG 59
S+ + QKK + F +FVG+L +S DVR+ F + I D R++ D T K KG G
Sbjct 84 STPSSQKKDTSNHFHVFVGDLSPEISTDDVRAAFAPFGK--ISDARVVKDLATGKSKGYG 141
Query 60 FVEFTNKEALKIAL-NYDGRTLNERKIRVEL 89
F+ F NK + A+ +G+ L R+IR
Sbjct 142 FISFINKWDAESAIQQMNGQWLGGRQIRTNW 172
> mmu:382035 Pabpn1l, Gm1108, Pabpnl1, ePABP2; poly(A)binding
protein nuclear 1-like; K14396 polyadenylate-binding protein
2
Length=273
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 41/73 (56%), Gaps = 2/73 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
+FVGN+ SAA++ ++F + I V +L DK + PKG ++EF + ++K A+
Sbjct 145 VFVGNVDYGGSAAELEAYFSPCGE--IHRVTILCDKFSGHPKGYAYIEFASHRSVKAAVG 202
Query 75 YDGRTLNERKIRV 87
D T R I+V
Sbjct 203 LDESTFRGRVIKV 215
> dre:394107 tial1, MGC66130, zgc:66130; TIA1 cytotoxic granule-associated
RNA binding protein-like 1; K13201 nucleolysin
TIA-1/TIAR
Length=370
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 46/88 (52%), Gaps = 3/88 (3%)
Query 3 SSEAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVE 62
SS+ + F +FVG+L ++ D+R+ F + I D R++ D T K KG GFV
Sbjct 86 SSQKKDTSNHFHVFVGDLSPEITTDDIRAAFAPFGK--ISDARVVKDMTTGKSKGYGFVS 143
Query 63 FTNK-EALKIALNYDGRTLNERKIRVEL 89
F NK +A ++ G+ L R+IR
Sbjct 144 FYNKLDAENAIVHMGGQWLGGRQIRTNW 171
> bbo:BBOV_I002350 19.m02195; RNA binding protein; K14396 polyadenylate-binding
protein 2
Length=163
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 41/73 (56%), Gaps = 2/73 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
++VGN+ + D++ FF Q I + ++ DK T KPKG ++EF +++A+ A+
Sbjct 44 IYVGNVDYSTKPHDLQEFFKASGQ--INRITIMVDKWTGKPKGYAYIEFASEDAVNNAVM 101
Query 75 YDGRTLNERKIRV 87
+ ER I+V
Sbjct 102 LNDCLFKERIIKV 114
> cel:R06C1.4 hypothetical protein
Length=84
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 44/79 (55%), Gaps = 3/79 (3%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
F ++VGN+P + ++ ++F + +VR++ D++T +P+G FVEF+ + + A
Sbjct 6 FSVYVGNVPYQGTEEEIGNYFAAVGH--VNNVRIVYDRETGRPRGFAFVEFSEEAGAQRA 63
Query 73 L-NYDGRTLNERKIRVELT 90
+ +G N R +RV
Sbjct 64 VEQLNGVAFNGRNLRVNYA 82
> hsa:7073 TIAL1, MGC33401, TCBP, TIAR; TIA1 cytotoxic granule-associated
RNA binding protein-like 1; K13201 nucleolysin TIA-1/TIAR
Length=392
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 46/88 (52%), Gaps = 3/88 (3%)
Query 3 SSEAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVE 62
SS+ + F +FVG+L ++ D++S F + I D R++ D T K KG GFV
Sbjct 104 SSQKKDTSNHFHVFVGDLSPEITTEDIKSAFAPFGK--ISDARVVKDMATGKSKGYGFVS 161
Query 63 FTNK-EALKIALNYDGRTLNERKIRVEL 89
F NK +A ++ G+ L R+IR
Sbjct 162 FYNKLDAENAIVHMGGQWLGGRQIRTNW 189
> dre:793165 tia1, MGC55520, fb73d10, wu:fb73d10, wu:fc02b04,
zgc:55520; cytotoxic granule-associated RNA binding protein
1; K13201 nucleolysin TIA-1/TIAR
Length=386
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 49/94 (52%), Gaps = 12/94 (12%)
Query 3 SSEAPQKK---SLFVLFVGNLPLTVSAADVRSFF---GRKLQLFIKDVRLLTDKQTKKPK 56
+S + QKK + F +FVG+L ++ D+R+ F GR I D R++ D T K K
Sbjct 83 TSPSSQKKDTSNHFHVFVGDLSPEITTDDIRAAFAPFGR-----ISDARVVKDMATGKSK 137
Query 57 GCGFVEFTNKEALKIAL-NYDGRTLNERKIRVEL 89
G GFV F NK + A+ G+ L R+IR
Sbjct 138 GYGFVSFFNKWDAENAIQQMGGQWLGGRQIRTNW 171
> mmu:21843 Tial1, 5330433G13Rik, AL033329, TIAR, mTIAR; Tia1
cytotoxic granule-associated RNA binding protein-like 1; K13201
nucleolysin TIA-1/TIAR
Length=392
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 46/88 (52%), Gaps = 3/88 (3%)
Query 3 SSEAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVE 62
SS+ + F +FVG+L ++ D++S F + I D R++ D T K KG GFV
Sbjct 104 SSQKKDTSNHFHVFVGDLSPEITTEDIKSAFAPFGK--ISDARVVKDMATGKSKGYGFVS 161
Query 63 FTNK-EALKIALNYDGRTLNERKIRVEL 89
F NK +A ++ G+ L R+IR
Sbjct 162 FYNKLDAENAIVHMGGQWLGGRQIRTNW 189
> ath:AT5G09880 RNA recognition motif (RRM)-containing protein;
K13091 RNA-binding protein 39
Length=527
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 43/74 (58%), Gaps = 2/74 (2%)
Query 5 EAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFT 64
EA ++ +F +PL + DV FF + + ++DVRL+ D+ +++ KG G++EF
Sbjct 160 EADPERDQRTVFAYQMPLKATERDVYEFFSKAGK--VRDVRLIMDRNSRRSKGVGYIEFY 217
Query 65 NKEALKIALNYDGR 78
+ ++ +A+ G+
Sbjct 218 DVMSVPMAIALSGQ 231
> ath:AT5G61030 GR-RBP3 (glycine-rich RNA-binding protein 3);
ATP binding / RNA binding
Length=309
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 43/76 (56%), Gaps = 3/76 (3%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
LF+G + ++ +R F + + + D R++ D++T + +G GFV FT+ EA A+
Sbjct 42 LFIGGMAYSMDEDSLREAFTKYGE--VVDTRVILDRETGRSRGFGFVTFTSSEAASSAIQ 99
Query 75 -YDGRTLNERKIRVEL 89
DGR L+ R ++V
Sbjct 100 ALDGRDLHGRVVKVNY 115
> tpv:TP03_0088 hypothetical protein; K14396 polyadenylate-binding
protein 2
Length=151
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 41/73 (56%), Gaps = 2/73 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
++VGN+ + +++ FF Q I + ++ DK T PKG +VEF+N++A+ A+
Sbjct 44 IYVGNVDYSTKPQELQEFFKSSGQ--INRITIMVDKYTGHPKGYAYVEFSNEDAVNNAIM 101
Query 75 YDGRTLNERKIRV 87
+ ER I+V
Sbjct 102 LNESLFKERIIKV 114
> ath:AT5G47620 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=431
Score = 45.1 bits (105), Expect = 7e-05, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 47/88 (53%), Gaps = 4/88 (4%)
Query 2 QSSEAPQKKSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFV 61
Q S P +FVG L +V+ A+ + +F + I DV ++ D +T++P+G GF+
Sbjct 97 QGSPGPSNSK--KIFVGGLASSVTEAEFKKYFAQFG--MITDVVVMYDHRTQRPRGFGFI 152
Query 62 EFTNKEALKIALNYDGRTLNERKIRVEL 89
+ ++EA+ L LN + + V+L
Sbjct 153 SYDSEEAVDKVLQKTFHELNGKMVEVKL 180
> ath:AT1G74230 GR-RBP5 (glycine-rich RNA-binding protein 5);
ATP binding / RNA binding
Length=289
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 41/75 (54%), Gaps = 2/75 (2%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIALN 74
+FVG + + +R F + + + D +++ D++T + +G FV FT+ E A+
Sbjct 36 IFVGGISYSTDEFGLREAFSKYGE--VVDAKIIVDRETGRSRGFAFVTFTSTEEASNAMQ 93
Query 75 YDGRTLNERKIRVEL 89
DG+ L+ R+IRV
Sbjct 94 LDGQDLHGRRIRVNY 108
> dre:368358 sb:cb657
Length=103
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 45/79 (56%), Gaps = 2/79 (2%)
Query 10 KSLFVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEAL 69
K +F +FV +P TV+ +V+ +FG+ Q +K L DK+T +G ++ FT +E L
Sbjct 10 KKVFEVFVTKVPWTVATKEVKEYFGQFGQ--VKRCLLPLDKETGFHRGFCWIGFTTEEGL 67
Query 70 KIALNYDGRTLNERKIRVE 88
+ AL D + K++V+
Sbjct 68 QNALQKDPHIIEGAKLQVQ 86
> ath:AT3G12640 RNA binding / nucleic acid binding / nucleotide
binding
Length=638
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 41/77 (53%), Gaps = 8/77 (10%)
Query 14 VLFVGNLPLTVSAADV-RSF--FGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALK 70
+FV N+ + + R F FG L+ FI +TD T +P G ++EFT KEA +
Sbjct 481 TIFVANVHFGATKDSLSRHFNKFGEVLKAFI-----VTDPATGQPSGSAYIEFTRKEAAE 535
Query 71 IALNYDGRTLNERKIRV 87
AL+ DG + R +++
Sbjct 536 NALSLDGTSFMSRILKI 552
> xla:100381100 tial1, tcbp, tiar; TIA1 cytotoxic granule-associated
RNA binding protein-like 1; K13201 nucleolysin TIA-1/TIAR
Length=385
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 42/81 (51%), Gaps = 9/81 (11%)
Query 13 FVLFVGNLPLTVSAADVRSFF---GRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEAL 69
F +FVG+L +S D+++ F GR I D R++ D T K KG GFV F NK
Sbjct 106 FHVFVGDLSPEISTDDIKAAFAPFGR-----ISDARVVKDMTTGKSKGYGFVSFFNKWDA 160
Query 70 KIAL-NYDGRTLNERKIRVEL 89
+ A+ G+ L R+IR
Sbjct 161 ENAIAQMGGQWLGGRQIRTNW 181
> bbo:BBOV_IV003970 23.m05815; RNA recognition motif containing
protein; K14838 nucleolar protein 15
Length=187
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 44/75 (58%), Gaps = 12/75 (16%)
Query 14 VLFVGNLPLTVSAADVRSFFGRKLQLF--IKDVRLLTDKQTKKPKGCGFVEFTNKEALKI 71
+++VGNLP ++ +++R +F + F +K +RL+ K+T +G F++F + E KI
Sbjct 11 IIYVGNLPKALNESNIRKYF----EQFGTVKKIRLMKSKKTGNSRGYCFLQFESNEIAKI 66
Query 72 AL----NY--DGRTL 80
A NY DGR L
Sbjct 67 AAEAMNNYFIDGRVL 81
> ath:AT3G46020 RNA-binding protein, putative
Length=102
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 44/76 (57%), Gaps = 3/76 (3%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIAL- 73
LFV L + +R F Q IK+ RL+ D +T++PKG GF+ F +++ + AL
Sbjct 9 LFVSRLSAYTTDQSLRQLFSPFGQ--IKEARLIRDSETQRPKGFGFITFDSEDDARKALK 66
Query 74 NYDGRTLNERKIRVEL 89
+ DG+ ++ R I VE+
Sbjct 67 SLDGKIVDGRLIFVEV 82
> sce:YBR212W NGR1, RBP1; Ngr1p
Length=672
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/76 (35%), Positives = 42/76 (55%), Gaps = 2/76 (2%)
Query 13 FVLFVGNLPLTVSAADVRSFFGRKLQLFIKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
F LFVG+L T + AD+ S F + + +K VR++TD T + GFV F +++ + A
Sbjct 192 FSLFVGDLSPTATEADLLSLFQTRFK-SVKTVRVMTDPLTGSSRCFGFVRFGDEDERRRA 250
Query 73 L-NYDGRTLNERKIRV 87
L G+ R +RV
Sbjct 251 LIEMSGKWFQGRALRV 266
> ath:AT3G07810 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=494
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 45/76 (59%), Gaps = 6/76 (7%)
Query 15 LFVGNLPLTVSAADVRSFFGRKLQLF--IKDVRLLTDKQTKKPKGCGFVEFTNKEALKIA 72
+FVG LP +V+ +D +++F + F DV ++ D T++P+G GF+ + ++EA++
Sbjct 110 IFVGGLPSSVTESDFKTYF----EQFGTTTDVVVMYDHNTQRPRGFGFITYDSEEAVEKV 165
Query 73 LNYDGRTLNERKIRVE 88
L LN + + V+
Sbjct 166 LLKTFHELNGKMVEVK 181
Lambda K H
0.315 0.133 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40