bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3342_orf2
Length=97
Score E
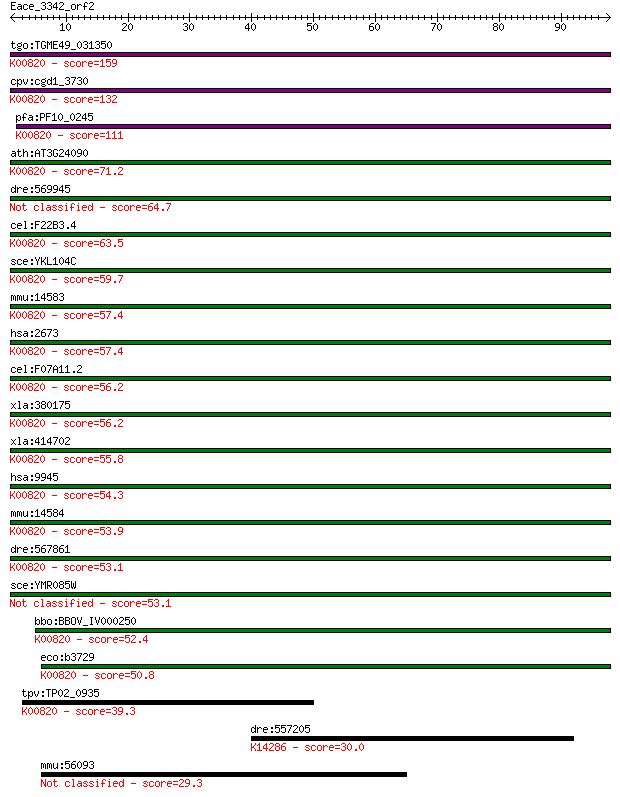
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotrans... 159 1e-39
cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferas... 132 3e-31
pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferas... 111 5e-25
ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (iso... 71.2 7e-13
dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase ... 64.7 7e-11
cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose... 63.5 1e-10
sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fru... 59.7 2e-09
mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,... 57.4 1e-08
hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutam... 57.4 1e-08
cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructos... 56.2 2e-08
xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosph... 56.2 2e-08
xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosph... 55.8 3e-08
hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphat... 54.3 9e-08
mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phospha... 53.9 1e-07
dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase ... 53.1 2e-07
sce:YMR085W Putative protein of unknown function; YMR085W and ... 53.1 2e-07
bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate... 52.4 4e-07
eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phos... 50.8 1e-06
tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferas... 39.3 0.003
dre:557205 MGC123007; zgc:123007; K14286 alanine-glyoxylate am... 30.0 1.8
mmu:56093 Pfpl, Epcs5, Epcs50; pore forming protein-like 29.3 2.9
> tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing), putative (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=603
Score = 159 bits (403), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 73/97 (75%), Positives = 86/97 (88%), Gaps = 0/97 (0%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGVY+NAGREVAVASTKAFTSQV VL+L+++WF QN ++ + DRCSAL+DA+HRLPVYA
Sbjct 380 CGVYVNAGREVAVASTKAFTSQVAVLSLIAAWFAQNQPTQAFPDRCSALMDAIHRLPVYA 439
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
GMTLN C+ IAE+LKDA+TLFVLGKGFGYPVALE
Sbjct 440 GMTLNCRALCQNIAERLKDAKTLFVLGKGFGYPVALE 476
> cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferase
(EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=696
Score = 132 bits (332), Expect = 3e-31, Method: Composition-based stats.
Identities = 60/97 (61%), Positives = 78/97 (80%), Gaps = 1/97 (1%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGVYLNAGREVAVASTKAF++QV VL+L+++WF QN R S RC LL+A+HR+P+
Sbjct 474 CGVYLNAGREVAVASTKAFSTQVLVLSLIAAWFAQN-RDSVISQRCQELLEAIHRVPISV 532
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
G++L A C++IAE +KD ++FVLGKG+GYPVALE
Sbjct 533 GVSLQAKDQCEQIAEMIKDNNSIFVLGKGYGYPVALE 569
> pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferase,
putative; K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=829
Score = 111 bits (278), Expect = 5e-25, Method: Composition-based stats.
Identities = 46/97 (47%), Positives = 75/97 (77%), Gaps = 1/97 (1%)
Query 2 GVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYAG 61
GVYLNAGREV VASTK FTS+V+VL L++ WF+Q+ ++ + S++ ++L+++LHRLP+Y G
Sbjct 606 GVYLNAGREVGVASTKCFTSEVSVLTLIALWFFQHKKNNQSSNKATSLINSLHRLPLYTG 665
Query 62 MTLNA-HGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
+T+ + CK ++EK K+ +++ ++G G YP+A E
Sbjct 666 VTIKSCENTCKTLSEKFKNTKSMLIIGNGLSYPIAQE 702
> ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (isomerizing)/
sugar binding / transaminase (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=695
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 56/97 (57%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ+ V+ +++ + S + R A++D L LP
Sbjct 472 CGVHINAGAEIGVASTKAYTSQIVVMVMLALAIGSDTISSQ--KRREAIIDGLLDLPYKV 529
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L K +A+ L D Q+L V G+G+ Y ALE
Sbjct 530 KEVLKLDDEMKDLAQLLIDEQSLLVFGRGYNYATALE 566
> dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase
2 (EC:2.6.1.16)
Length=681
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 38/97 (39%), Positives = 53/97 (54%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG EV VASTKA+TSQ L + ++ S E R ++ L++LP
Sbjct 460 CGVHINAGPEVGVASTKAYTSQFVSLIMFGLMMCEDRLSLE--PRRQEIISGLNQLPELI 517
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L K IAE+L ++L V+G+GF Y LE
Sbjct 518 KKVLAQDDNIKTIAEELHHQRSLLVMGRGFNYATCLE 554
> cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=710
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 33/97 (34%), Positives = 58/97 (59%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CG+++NAG E+ VASTKA+TSQ+ L L + ++ S+ R + ++DAL+ LP+
Sbjct 488 CGIHINAGPEIGVASTKAYTSQIVSLLLFALTISEDRISK--MKRRAEIIDALNNLPILI 545
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +IAE++ ++L ++G+G + LE
Sbjct 546 RDVLDLDDQVLKIAEQIYKDKSLLIMGRGLNFATCLE 582
> sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=717
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 56/100 (56%), Gaps = 7/100 (7%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYS--DRCSALLDALHRLPV 58
CGV++NAG E+ VASTKA+TSQ L + F +L + S DR ++ L +P
Sbjct 491 CGVHINAGPEIGVASTKAYTSQYIALVM----FALSLSDDRVSKIDRRIEIIQGLKLIPG 546
Query 59 YAGMTLNAHGACKRI-AEKLKDAQTLFVLGKGFGYPVALE 97
L K++ A +LKD ++L +LG+G+ + ALE
Sbjct 547 QIKQVLKLEPRIKKLCATELKDQKSLLLLGRGYQFAAALE 586
> mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,
GFAT1, GFAT1m, Gfpt; glutamine fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + + S + +R ++ L RLP
Sbjct 460 CGVHINAGPEIGVASTKAYTSQFVSLVMFALMMCDDRISMQ--ERRKEIMLGLKRLPDLI 517
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +++A +L +++ ++G+G+ Y LE
Sbjct 518 KEVLSMDDEIQKLATELYHQKSVLIMGRGYHYATCLE 554
> hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutamine--fructose-6-phosphate
transaminase 1 (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=681
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + + S + +R ++ L RLP
Sbjct 460 CGVHINAGPEIGVASTKAYTSQFVSLVMFALMMCDDRISMQ--ERRKEIMLGLKRLPDLI 517
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +++A +L +++ ++G+G+ Y LE
Sbjct 518 KEVLSMDDEIQKLATELYHQKSVLIMGRGYHYATCLE 554
> cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=712
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 33/99 (33%), Positives = 53/99 (53%), Gaps = 6/99 (6%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYS--DRCSALLDALHRLPV 58
CG+++NAG E+ VASTKA+TSQ+ L + F L + S R ++DAL+ LP
Sbjct 490 CGIHINAGPEIGVASTKAYTSQILSLLM----FALTLSDDRISMAKRREEIIDALNDLPE 545
Query 59 YAGMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L IA+++ ++L ++G+G + LE
Sbjct 546 LIREVLQLDEKVLDIAKQIYKEKSLLIMGRGLNFATCLE 584
> xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 55/97 (56%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + + S + +R +++AL LP
Sbjct 460 CGVHINAGPEIGVASTKAYTSQFVSLIMFALMMCDDRISIQ--ERRKQIMNALQILPDNI 517
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +++A +L +++ ++G+G+ Y +E
Sbjct 518 KEVLSLDDEIQKLASELYQQKSVLIMGRGYHYSTCME 554
> xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosphate
transaminase 1; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + + S + +R +++ L LP
Sbjct 460 CGVHINAGPEIGVASTKAYTSQFVSLIMFALMMCDDRISMQ--ERRKQIINGLKILPDNI 517
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +++A +L +++ ++G+GF Y +E
Sbjct 518 KEVLSLDDEIQKLASELYQQKSVLIMGRGFHYATCME 554
> hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 32/97 (32%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + ++ S + +R ++ L LP
Sbjct 461 CGVHINAGPEIGVASTKAYTSQFISLVMFGLMMSEDRISLQ--NRRQEIIRGLRSLPELI 518
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +A +L ++L V+G+G+ Y LE
Sbjct 519 KEVLSLEEKIHDLALELYTQRSLLVMGRGYNYATCLE 555
> mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/97 (32%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + ++ S + +R ++ L LP
Sbjct 461 CGVHINAGPEIGVASTKAYTSQFISLVMFGLMMSEDRISLQ--NRRQEIIRGLRSLPELI 518
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L+ +A +L ++L V+G+G+ Y LE
Sbjct 519 KEVLSLDEKIHDLALELYTQRSLLVMGRGYNYATCLE 555
> dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV++NAG E+ VASTKA+TSQ L + + + S + R ++ L LP
Sbjct 461 CGVHINAGPEIGVASTKAYTSQFVALVMFALLMCDDRISVQ--PRRKEIIQGLRILPDLI 518
Query 61 GMTLNAHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
L +++A +L +++ ++G+G+ Y LE
Sbjct 519 KEVLTLDEEIQKLAAELYPQKSVLIMGRGYHYATCLE 555
> sce:YMR085W Putative protein of unknown function; YMR085W and
adjacent ORF YMR084W are merged in related strains
Length=432
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 3/98 (3%)
Query 1 CGVYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYA 60
CGV+ N G E +A+TK++TSQ L +++ W ++L S+ +R ++ AL +P
Sbjct 206 CGVHTNTGPEKGIATTKSYTSQYIALVMIALWMSEDLVSK--IERRKEIIQALTIVPSQI 263
Query 61 GMTLNAHGACKRIAE-KLKDAQTLFVLGKGFGYPVALE 97
L + + KLK T +LG+G+ + ALE
Sbjct 264 KEVLELEPLIIELCDKKLKQHDTFLLLGRGYQFASALE 301
> bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=723
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 53/97 (54%), Gaps = 7/97 (7%)
Query 5 LNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYAGMTL 64
+N GRE+++ASTK FT Q+ +L + + Q E++ + L + + + G+ L
Sbjct 505 VNIGREISLASTKTFTVQIMLLMALIGYIVQAQDLEKHHEE---FLRQMKKSIIGYGLAL 561
Query 65 N----AHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
C+R+AE+L + +++V+G G GY +A E
Sbjct 562 KKVLCTEDICQRVAERLCNVDSMYVIGSGEGYAIAQE 598
> eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=609
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 33/92 (35%), Positives = 48/92 (52%), Gaps = 2/92 (2%)
Query 6 NAGREVAVASTKAFTSQVTVLALVSSWFWQNLRSEEYSDRCSALLDALHRLPVYAGMTLN 65
NAG E+ VASTKAFT+Q+TVL ++ + L+ + S ++ L LP L+
Sbjct 393 NAGTEIGVASTKAFTTQLTVLLMLVAKL-SRLKGLDASIE-HDIVHGLQALPSRIEQMLS 450
Query 66 AHGACKRIAEKLKDAQTLFVLGKGFGYPVALE 97
+ +AE D LG+G YP+ALE
Sbjct 451 QDKRIEALAEDFSDKHHALFLGRGDQYPIALE 482
> tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferase;
K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=810
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 33/52 (63%), Gaps = 5/52 (9%)
Query 3 VYLNAGREVAVASTKAFTSQVTVLALVSSWFWQNLRS-----EEYSDRCSAL 49
++L GRE +V STKAFT+QVTVL ++S + N ++ +D CS++
Sbjct 534 IHLRIGREKSVPSTKAFTAQVTVLLILSLYIINNYDITIDPLDDSTDYCSSV 585
> dre:557205 MGC123007; zgc:123007; K14286 alanine-glyoxylate
aminotransferase 2-like [EC:2.6.1.-]
Length=447
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 31/67 (46%), Gaps = 15/67 (22%)
Query 40 EEYSDRCSALLDALHR------------LPVYAGMTLNAHGACKRIAEKLKDAQTLFV-- 85
+ Y+D +L++ H+ LP G + G CKR+AE + +A ++V
Sbjct 187 QAYADTVKSLIEEAHKKGRKISSFFAESLPSVGGQIIFPTGYCKRVAEYVHEAGGVYVAD 246
Query 86 -LGKGFG 91
+ GFG
Sbjct 247 EIQTGFG 253
> mmu:56093 Pfpl, Epcs5, Epcs50; pore forming protein-like
Length=702
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 17/61 (27%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 6 NAGREVAVASTKAFT-SQVTVLALVSSWFWQNLRSEEY-SDRCSALLDALHRLPVYAGMT 63
N G AVA++ FT ++V + + + +QN + Y +R + + ++ +P Y GMT
Sbjct 229 NKGNRAAVAASAGFTFAKVVNFKIETGFEYQNNLATGYLQNRTGSRVQSIGGIPFYPGMT 288
Query 64 L 64
+
Sbjct 289 I 289
Lambda K H
0.322 0.133 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40