bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3407_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
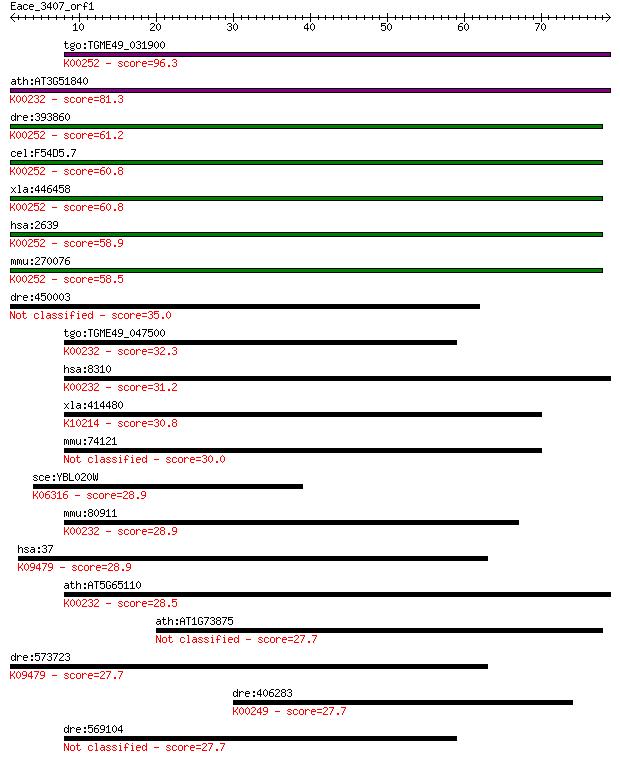
tgo:TGME49_031900 acyl-CoA dehydrogenase, putative (EC:1.3.99.... 96.3 2e-20
ath:AT3G51840 ACX4; ACX4 (ACYL-COA OXIDASE 4); acyl-CoA oxidas... 81.3 8e-16
dre:393860 gcdh, MGC77704, zgc:56505, zgc:77704; glutaryl-Coen... 61.2 7e-10
cel:F54D5.7 hypothetical protein; K00252 glutaryl-CoA dehydrog... 60.8 1e-09
xla:446458 gcdh, MGC78897; glutaryl-CoA dehydrogenase (EC:1.3.... 60.8 1e-09
hsa:2639 GCDH, ACAD5, GCD; glutaryl-CoA dehydrogenase (EC:1.3.... 58.9 4e-09
mmu:270076 Gcdh, 9030411L18, AI266902, D17825; glutaryl-Coenzy... 58.5 5e-09
dre:450003 gcdhl, zgc:103477; glutaryl-Coenzyme A dehydrogenas... 35.0 0.053
tgo:TGME49_047500 acyl-coenzyme A oxidase, putative (EC:1.9.3.... 32.3 0.35
hsa:8310 ACOX3; acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6); K... 31.2 0.97
xla:414480 acox2, MGC83074; acyl-CoA oxidase 2, branched chain... 30.8 0.98
mmu:74121 Acoxl, 1200014P05Rik, AV025673; acyl-Coenzyme A oxid... 30.0 2.2
sce:YBL020W RFT1; Essential integral membrane protein that is ... 28.9 3.8
mmu:80911 Acox3, BI685180, EST-s59, PCOX; acyl-Coenzyme A oxid... 28.9 4.2
hsa:37 ACADVL, ACAD6, LCACD, VLCAD; acyl-CoA dehydrogenase, ve... 28.9 4.3
ath:AT5G65110 ACX2; ACX2 (ACYL-COA OXIDASE 2); acyl-CoA oxidas... 28.5 6.2
ath:AT1G73875 endonuclease/exonuclease/phosphatase family protein 27.7 8.5
dre:573723 acadvl, MGC174519, MGC64067, fb52d04, wu:fb52d04, w... 27.7 9.0
dre:406283 acadm, fb53e01, wu:fb53e01, zgc:111905, zgc:56101, ... 27.7 9.7
dre:569104 si:ch211-205h19.1 27.7 10.0
> tgo:TGME49_031900 acyl-CoA dehydrogenase, putative (EC:1.3.99.7);
K00252 glutaryl-CoA dehydrogenase [EC:1.3.99.7]
Length=454
Score = 96.3 bits (238), Expect = 2e-20, Method: Composition-based stats.
Identities = 50/71 (70%), Positives = 54/71 (76%), Gaps = 0/71 (0%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIA 67
A I+L K T S +RE I LCRE +GGNGI+ AK ADIEALYTYEGTYDINMLIA
Sbjct 384 AHIALAKATCSRVAREGIALCREVLGGNGIVTDFGVAKFHADIEALYTYEGTYDINMLIA 443
Query 68 GRAVTGLSAFK 78
GRAVTG SAFK
Sbjct 444 GRAVTGRSAFK 454
> ath:AT3G51840 ACX4; ACX4 (ACYL-COA OXIDASE 4); acyl-CoA oxidase/
oxidoreductase (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=436
Score = 81.3 bits (199), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 40/78 (51%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
++G ++P SL K S +RE L RE +GGNGIL AKA D+E +YTYEGTY
Sbjct 352 ETGQMTPGQASLGKAWISSKARETASLGRELLGGNGILADFLVAKAFCDLEPIYTYEGTY 411
Query 61 DINMLIAGRAVTGLSAFK 78
DIN L+ GR VTG+++FK
Sbjct 412 DINTLVTGREVTGIASFK 429
> dre:393860 gcdh, MGC77704, zgc:56505, zgc:77704; glutaryl-Coenzyme
A dehydrogenase (EC:1.3.99.7); K00252 glutaryl-CoA dehydrogenase
[EC:1.3.99.7]
Length=441
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
D +P +ISL+K + + + + R+ +GGNGI + + ++EA+ TYEGT+
Sbjct 361 DQKKHTPEMISLLKRNSCGKALDIARQARDMLGGNGIADEYHIIRHVMNLEAVNTYEGTH 420
Query 61 DINMLIAGRAVTGLSAF 77
DI+ LI GRA+TGL +F
Sbjct 421 DIHALILGRAITGLQSF 437
> cel:F54D5.7 hypothetical protein; K00252 glutaryl-CoA dehydrogenase
[EC:1.3.99.7]
Length=409
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
D G + IS+IK + + E + R+ +GGNGI+ + + ++E + TYEGT+
Sbjct 332 DEGKVQSEQISIIKRNSCGKALEVARKARDMLGGNGIVDEYHIMRHMVNLETVNTYEGTH 391
Query 61 DINMLIAGRAVTGLSAF 77
D++ LI GRA+TGL+ F
Sbjct 392 DVHALILGRAITGLNGF 408
> xla:446458 gcdh, MGC78897; glutaryl-CoA dehydrogenase (EC:1.3.99.7);
K00252 glutaryl-CoA dehydrogenase [EC:1.3.99.7]
Length=440
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
D +P +IS++K + + + + R+ +GGNGI + + ++EA+ TYEGT+
Sbjct 360 DEKRAAPEMISMLKRNSCGKALDIARQARDMLGGNGISDEYHIIRHVMNLEAVNTYEGTH 419
Query 61 DINMLIAGRAVTGLSAF 77
D++ LI GRA+TGL AF
Sbjct 420 DVHALILGRAITGLQAF 436
> hsa:2639 GCDH, ACAD5, GCD; glutaryl-CoA dehydrogenase (EC:1.3.99.7);
K00252 glutaryl-CoA dehydrogenase [EC:1.3.99.7]
Length=438
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 46/77 (59%), Gaps = 0/77 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
D +P ++SL+K + + + R+ +GGNGI + ++EA+ TYEGT+
Sbjct 358 DQDKAAPEMVSLLKRNNCGKALDIARQARDMLGGNGISDEYHVIRHAMNLEAVNTYEGTH 417
Query 61 DINMLIAGRAVTGLSAF 77
DI+ LI GRA+TG+ AF
Sbjct 418 DIHALILGRAITGIQAF 434
> mmu:270076 Gcdh, 9030411L18, AI266902, D17825; glutaryl-Coenzyme
A dehydrogenase (EC:1.3.99.7); K00252 glutaryl-CoA dehydrogenase
[EC:1.3.99.7]
Length=438
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 46/77 (59%), Gaps = 0/77 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
D +P ++S++K + + + R+ +GGNGI + ++EA+ TYEGT+
Sbjct 358 DQDKATPEMVSMLKRNNCGKALDIARQARDILGGNGISDEYHVIRHAMNLEAVNTYEGTH 417
Query 61 DINMLIAGRAVTGLSAF 77
DI+ LI GRA+TG+ AF
Sbjct 418 DIHALILGRAITGIQAF 434
> dre:450003 gcdhl, zgc:103477; glutaryl-Coenzyme A dehydrogenase,
like
Length=427
Score = 35.0 bits (79), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 33/61 (54%), Gaps = 0/61 (0%)
Query 1 DSGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTY 60
D +P +ISL+K + + + + R+ +GGNGI + + ++EA+ TYEG
Sbjct 360 DDKKAAPEMISLLKRNSCGKALDIARQARDMLGGNGIADEYHIIRHVLNLEAVNTYEGQR 419
Query 61 D 61
+
Sbjct 420 E 420
> tgo:TGME49_047500 acyl-coenzyme A oxidase, putative (EC:1.9.3.1);
K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=696
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEG 58
A+ S +K SL + ++ CR+A GG+G LL + + TYEG
Sbjct 393 AVTSCLKAMMSLSVADGMEKCRKACGGHGFLLASGIPIQFVNFVPVATYEG 443
> hsa:8310 ACOX3; acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6);
K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=624
Score = 31.2 bits (69), Expect = 0.97, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 0/71 (0%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIA 67
AL S K S +++ I+ CREA GG+G L + D + TYEG +I +
Sbjct 402 ALASASKPLASWTTQQGIQECREACGGHGYLAMNRLGVLRDDNDPNCTYEGDNNILLQQT 461
Query 68 GRAVTGLSAFK 78
+ GL A +
Sbjct 462 SNYLLGLLAHQ 472
> xla:414480 acox2, MGC83074; acyl-CoA oxidase 2, branched chain;
K10214 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestanoyl-CoA
24-hydroxylase [EC:1.17.99.3]
Length=670
Score = 30.8 bits (68), Expect = 0.98, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 0/62 (0%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIA 67
AL+S +K + I++CR+A GG+G L + A TYEG + L A
Sbjct 383 ALVSGMKAYATETCSNGIEVCRKACGGHGYSLFSGLPSLYTKVTASCTYEGENTVLHLQA 442
Query 68 GR 69
R
Sbjct 443 AR 444
> mmu:74121 Acoxl, 1200014P05Rik, AV025673; acyl-Coenzyme A oxidase-like
Length=632
Score = 30.0 bits (66), Expect = 2.2, Method: Composition-based stats.
Identities = 14/62 (22%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIA 67
AL++ +K ++ + ++ CRE GG G ++ + + D + T+EG + + +
Sbjct 352 ALMAGLKAYSTWETVSCLQDCRECTGGMGYMMETRISDLKCDTDVFVTFEGDNVVMLQVV 411
Query 68 GR 69
R
Sbjct 412 AR 413
> sce:YBL020W RFT1; Essential integral membrane protein that is
required for translocation of Man5GlcNac2-PP-Dol from the
cytoplasmic side to the lumenal side of the ER membrane but
is not the flippase; mutation is suppressed by expression of
human p53 protein; K06316 oligosaccharidyl-lipid flippase family
Length=574
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 25/43 (58%), Gaps = 8/43 (18%)
Query 4 FLSP------ALISLIKGTTSLWSREAIKLC--REAMGGNGIL 38
FLSP A + I+GT +SR+AI+L R + GNGI+
Sbjct 47 FLSPRIFGITAFLEFIQGTVLFFSRDAIRLSTLRISDSGNGII 89
> mmu:80911 Acox3, BI685180, EST-s59, PCOX; acyl-Coenzyme A oxidase
3, pristanoyl (EC:1.3.3.6); K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=700
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLI 66
AL S K S ++ I+ CREA GG+G L + D + TYEG D N+L+
Sbjct 402 ALASAGKPLASWTAQRGIQECREACGGHGYLAMNRFGDLRNDNDPNCTYEG--DNNVLL 458
> hsa:37 ACADVL, ACAD6, LCACD, VLCAD; acyl-CoA dehydrogenase,
very long chain (EC:1.3.99.-); K09479 very long chain acyl-CoA
dehydrogenase [EC:1.3.99.-]
Length=655
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 2 SGFLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYD 61
+ F A IS I G+ + W + C + MGG G + + L D+ +EGT D
Sbjct 409 TDFQIEAAISKIFGSEAAW--KVTDECIQIMGGMGFMKEPGVERVLRDLRIFRIFEGTND 466
Query 62 I 62
I
Sbjct 467 I 467
> ath:AT5G65110 ACX2; ACX2 (ACYL-COA OXIDASE 2); acyl-CoA oxidase;
K00232 acyl-CoA oxidase [EC:1.3.3.6]
Length=646
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 2/71 (2%)
Query 8 ALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIA 67
AL + +K + ++ +A+ +CREA GG+G + D + T+EG D +L+
Sbjct 426 ALSAGLKSYVTSYTAKALSVCREACGGHGYAAVNRFGSLRNDHDIFQTFEG--DNTVLLQ 483
Query 68 GRAVTGLSAFK 78
A L +K
Sbjct 484 QVAADLLKRYK 494
> ath:AT1G73875 endonuclease/exonuclease/phosphatase family protein
Length=454
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 24/58 (41%), Gaps = 0/58 (0%)
Query 20 WSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIAGRAVTGLSAF 77
WSR +C+E N +L LQ D++ L G ++ G A G + F
Sbjct 130 WSRRKHLICKEISRYNASILCLQEVDRFDDLDVLLKNRGFRGVHKSRTGEASDGCAIF 187
> dre:573723 acadvl, MGC174519, MGC64067, fb52d04, wu:fb52d04,
wu:fc75e01, zgc:64067; acyl-Coenzyme A dehydrogenase, very
long chain; K09479 very long chain acyl-CoA dehydrogenase [EC:1.3.99.-]
Length=659
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 28/65 (43%), Gaps = 5/65 (7%)
Query 1 DSG---FLSPALISLIKGTTSLWSREAIKLCREAMGGNGILLGLQTAKALADIEALYTYE 57
DSG F A IS I + + W C + MGG G + + L D+ +E
Sbjct 410 DSGATEFQIEAAISKIFASEAAWL--VTDECIQVMGGMGFMKDAGVERVLRDLRIFRIFE 467
Query 58 GTYDI 62
GT DI
Sbjct 468 GTNDI 472
> dre:406283 acadm, fb53e01, wu:fb53e01, zgc:111905, zgc:56101,
zgc:76911; acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight
chain (EC:1.3.99.3); K00249 acyl-CoA dehydrogenase [EC:1.3.99.3]
Length=424
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 19/44 (43%), Gaps = 0/44 (0%)
Query 30 EAMGGNGILLGLQTAKALADIEALYTYEGTYDINMLIAGRAVTG 73
+ GGNG K + D + YEGT I LI R V G
Sbjct 377 QIFGGNGFNSEYPVEKLMRDAKIYQIYEGTAQIQRLIISREVLG 420
> dre:569104 si:ch211-205h19.1
Length=652
Score = 27.7 bits (60), Expect = 10.0, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 27/54 (50%), Gaps = 3/54 (5%)
Query 8 ALISLIKG--TTSLWSREA-IKLCREAMGGNGILLGLQTAKALADIEALYTYEG 58
AL +L+ G S W+ A ++ CRE GG G ++ + D + T+EG
Sbjct 373 ALQALVAGLKAYSTWANVACLQDCRECTGGMGFMMENRIPSLKCDSDVFVTFEG 426
Lambda K H
0.319 0.136 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40