bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0045_orf1
Length=189
Score E
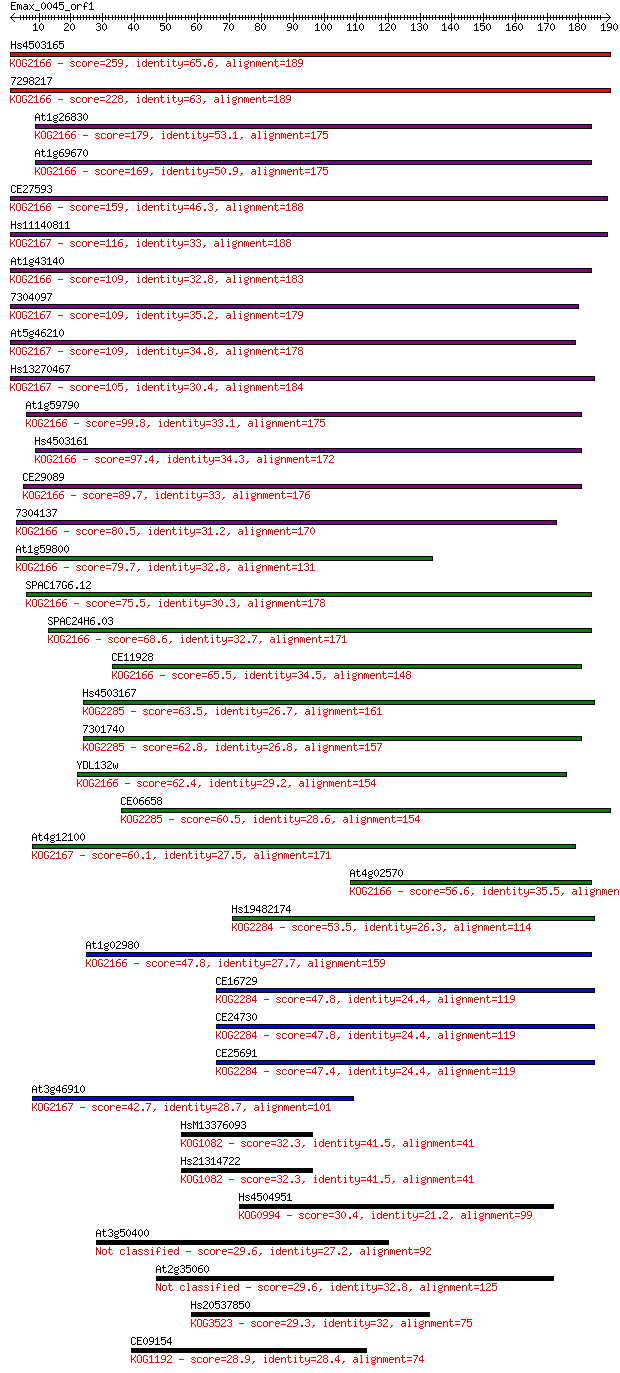
Sequences producing significant alignments: (Bits) Value
Hs4503165 259 2e-69
7298217 228 6e-60
At1g26830 179 4e-45
At1g69670 169 3e-42
CE27593 159 3e-39
Hs11140811 116 2e-26
At1g43140 109 3e-24
7304097 109 3e-24
At5g46210 109 4e-24
Hs13270467 105 5e-23
At1g59790 99.8 3e-21
Hs4503161 97.4 2e-20
CE29089 89.7 3e-18
7304137 80.5 2e-15
At1g59800 79.7 3e-15
SPAC17G6.12 75.5 6e-14
SPAC24H6.03 68.6 7e-12
CE11928 65.5 6e-11
Hs4503167 63.5 2e-10
7301740 62.8 4e-10
YDL132w 62.4 6e-10
CE06658 60.5 2e-09
At4g12100 60.1 3e-09
At4g02570 56.6 3e-08
Hs19482174 53.5 2e-07
At1g02980 47.8 1e-05
CE16729 47.8 1e-05
CE24730 47.8 1e-05
CE25691 47.4 2e-05
At3g46910 42.7 4e-04
HsM13376093 32.3 0.57
Hs21314722 32.3 0.57
Hs4504951 30.4 1.9
At3g50400 29.6 3.8
At2g35060 29.6 3.9
Hs20537850 29.3 4.4
CE09154 28.9 6.2
> Hs4503165
Length=768
Score = 259 bits (662), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 124/189 (65%), Positives = 154/189 (81%), Gaps = 0/189 (0%)
Query 1 VIQNNVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHML 60
V QNNV++V+NLGLI+FRD VVR+G IRDH+R+TLL+++ RER GE++DR A++N C ML
Sbjct 131 VQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEVVDRGAIRNACQML 190
Query 61 IQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACH 120
+ LG+ R+VYE+DFE FL+ SAEF+++ESQKFL N+A YIKKVE RINEE ER H
Sbjct 191 MILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKVEARINEEIERVMH 250
Query 121 YLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGL 180
LD STE+ IVKVVE ELIS HMKTIVEMENSG+VHML+N K EDL MYKLF RV +GL
Sbjct 251 CLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLGCMYKLFSRVPNGL 310
Query 181 KTIIACVST 189
KT+ C+S+
Sbjct 311 KTMCECMSS 319
> 7298217
Length=773
Score = 228 bits (581), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 119/190 (62%), Positives = 139/190 (73%), Gaps = 1/190 (0%)
Query 1 VIQNNVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHML 60
V Q VD+V+NLGLILFRD VVR+ I+ +RE LL +V+ ER+GE I+ LA+KN C ML
Sbjct 129 VQQREVDNVYNLGLILFRDQVVRYSEIQKALREKLLGMVMEERHGEAINHLAIKNACSML 188
Query 61 IQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACH 120
I LGINSRTVYE+DFEK FL QSA FYK ESQ FL N A YIKKVE RI EE+ RA
Sbjct 189 ITLGINSRTVYEEDFEKPFLAQSAAFYKFESQNFLAENNAGVYIKKVEARITEESSRAAL 248
Query 121 YLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRV-DSG 179
YLD TE IV+VVE ELI HM+ IVEMENSGVV+M++N K EDLA YKLF R+ + G
Sbjct 249 YLDKDTEPRIVRVVEEELIKKHMRPIVEMENSGVVYMIKNSKTEDLACTYKLFSRLKEEG 308
Query 180 LKTIIACVST 189
LK I +S
Sbjct 309 LKVIADTMSA 318
> At1g26830
Length=732
Score = 179 bits (453), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 93/175 (53%), Positives = 123/175 (70%), Gaps = 3/175 (1%)
Query 9 VFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSR 68
V +GL L+RD+VV F I + TLL+LV +ER GE+IDR ++N M + LG
Sbjct 133 VHPMGLNLWRDNVVHFTKIHTRLLNTLLDLVQKERIGEVIDRGLMRNVIKMFMDLG---E 189
Query 69 TVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQ 128
+VY++DFEK FL S+EFYK+ESQ+F+E DY+KK E+R+ EE ER HYLDA +E+
Sbjct 190 SVYQEDFEKPFLDASSEFYKVESQEFIESCDCGDYLKKSEKRLTEEIERVAHYLDAKSEE 249
Query 129 LIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTI 183
I VVE E+I+NHM+ +V MENSG+V+ML N K EDL RMY LF RV +GL T+
Sbjct 250 KITSVVEKEMIANHMQRLVHMENSGLVNMLLNDKYEDLGRMYNLFRRVTNGLVTV 304
> At1g69670
Length=732
Score = 169 bits (428), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 89/175 (50%), Positives = 122/175 (69%), Gaps = 3/175 (1%)
Query 9 VFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSR 68
V LGL L+RD+VV I+ + TLL+LV +ER GE+IDR+ ++N M + LG
Sbjct 133 VHELGLHLWRDNVVYSSKIQTRLLNTLLDLVHKERTGEVIDRVLMRNVIKMFMDLG---E 189
Query 69 TVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQ 128
+VY+DDFEK FL+ SAEFYK+ES +F+E +Y+KK E+ + EE ER +YLDA +E
Sbjct 190 SVYQDDFEKPFLEASAEFYKVESMEFIESCDCGEYLKKAEKPLVEEVERVVNYLDAKSEA 249
Query 129 LIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTI 183
I VVE E+I+NH++ +V MENSG+V+ML N K ED+ RMY LF RV +GL T+
Sbjct 250 KITSVVEREMIANHVQRLVHMENSGLVNMLLNDKYEDMGRMYSLFRRVANGLVTV 304
> CE27593
Length=777
Score = 159 bits (403), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 87/197 (44%), Positives = 132/197 (67%), Gaps = 9/197 (4%)
Query 1 VIQNN-VDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHM 59
V QNN V V+NLGL +R ++R I D IR+ LLEL+ +R I+ +KN C M
Sbjct 128 VAQNNHVLPVYNLGLDAYRTEILRQNGIGDRIRDALLELIKLDRKSNQINWHGIKNACDM 187
Query 60 LIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEG-NTACDYIKKVEQRINEEAERA 118
LI LGI+SRTVYED+FE+ L++++++Y+ + +L G N AC Y+ +VE +++EA RA
Sbjct 188 LISLGIDSRTVYEDEFERPLLKETSDYYRDVCKNWLSGDNDACFYLAQVEIAMHDEASRA 247
Query 119 CHYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRV-D 177
YLD TE I++V++ +++ H++TIV M+N GV MLE++KIEDL R++++F R+ D
Sbjct 248 SRYLDKMTEAKILQVMDDVMVAEHIQTIVYMQNGGVKFMLEHKKIEDLTRIFRIFKRIGD 307
Query 178 S------GLKTIIACVS 188
S GLK ++ VS
Sbjct 308 SVTVPGGGLKALLKAVS 324
> Hs11140811
Length=659
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 62/189 (32%), Positives = 113/189 (59%), Gaps = 9/189 (4%)
Query 1 VIQNN-VDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHM 59
V+QN+ + S++++GL LFR H++ +++ + +L L+ RER+GE +DR +++ M
Sbjct 61 VLQNSTLPSIWDMGLELFRTHIISDKMVQSKTIDGILLLIERERSGEAVDRSLLRSLLGM 120
Query 60 LIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC 119
L L VY+D FE FL+++ Y E Q+ ++ +Y+ V +R+ EE +R
Sbjct 121 LSDL-----QVYKDSFELKFLEETNCLYAAEGQRLMQEREVPEYLNHVSKRLEEEGDRVI 175
Query 120 HYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSG 179
YLD ST++ ++ VE +L+ H+ I++ G+ H+L+ ++ DLA+MY+LF RV G
Sbjct 176 TYLDHSTQKPLIACVEKQLLGEHLTAILQ---KGLDHLLDENRVPDLAQMYQLFSRVRGG 232
Query 180 LKTIIACVS 188
+ ++ S
Sbjct 233 QQALLQHWS 241
> At1g43140
Length=711
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 60/183 (32%), Positives = 107/183 (58%), Gaps = 2/183 (1%)
Query 1 VIQNNVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHML 60
V + + ++ ++G F D V + I+ ++ LL L+ +ER GE IDR VKN +
Sbjct 119 VARRGLPTLNDVGFTSFHDLV--YQEIQSEAKDVLLALIHKEREGEQIDRTLVKNVIDVY 176
Query 61 IQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACH 120
G+ +YE+DFE LQ +A +Y ++ ++ + ++ DY+ K E+ + E ER +
Sbjct 177 CGNGVGQMVIYEEDFESFLLQDTASYYSRKASRWSQEDSCPDYMLKAEECLKLEKERVTN 236
Query 121 YLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGL 180
YL ++TE +V+ V+ EL+ K ++E E+SG + +L + K+ DL+RMY+L+ + GL
Sbjct 237 YLHSTTEPKLVEKVQNELLVVVAKQLIENEHSGCLALLRDDKMGDLSRMYRLYRLIPQGL 296
Query 181 KTI 183
+ I
Sbjct 297 EPI 299
> 7304097
Length=773
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 63/180 (35%), Positives = 107/180 (59%), Gaps = 9/180 (5%)
Query 1 VIQNN-VDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHM 59
V+QN+ V S++++GL LFR H + +++ + LL L+ +ER G +DR +K+ M
Sbjct 213 VLQNSTVHSIWDMGLDLFRIHFAQNSVVQKRTVDGLLTLIEKERQGSTVDRGLLKSLVRM 272
Query 60 LIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC 119
L L I Y FE+ FL + + YK ESQ+ ++ +Y++ V +R+ EE ER
Sbjct 273 LCDLQI-----YTSSFEEKFLDATNQLYKAESQRKMQELEVPEYLQHVNKRLAEENERLR 327
Query 120 HYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSG 179
HYLD+ST+ ++ VE EL++ H+ +I++ G+ +LE+ ++ DL +Y L RV +G
Sbjct 328 HYLDSSTKHPLIYNVEKELLAEHLTSILQ---KGLDSLLEDNRLSDLTLLYGLLSRVKNG 384
> At5g46210
Length=617
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 62/179 (34%), Positives = 103/179 (57%), Gaps = 9/179 (5%)
Query 1 VIQN-NVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHM 59
VIQN NV S++ +GL LFR H+ + + LL ++ +ER E ++R + + M
Sbjct 16 VIQNPNVRSLWEMGLQLFRKHLSLAPEVEQRTVKGLLSMIEKERLAEAVNRTLLSHLLKM 75
Query 60 LIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC 119
LGI Y + FEK FL+ ++EFY E K+++ + +Y+K VE R++EE ER
Sbjct 76 FTALGI-----YMESFEKPFLEGTSEFYAAEGMKYMQQSDVPEYLKHVEGRLHEENERCI 130
Query 120 HYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDS 178
Y+DA T + ++ VE +L+ H+ ++E G +++ ++ EDL RM LF RV++
Sbjct 131 LYIDAVTRKPLITTVERQLLERHILVVLE---KGFTTLMDGRRTEDLQRMQTLFSRVNA 186
> Hs13270467
Length=717
Score = 105 bits (262), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 56/185 (30%), Positives = 112/185 (60%), Gaps = 9/185 (4%)
Query 1 VIQNN-VDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHM 59
V+QN+ + S++++GL LFR H++ +++ + +L L+ RERNGE A+ +
Sbjct 119 VLQNSMLPSIWDMGLELFRAHIISDQKVQNKTIDGILLLIERERNGE-----AIDRSLLR 173
Query 60 LIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC 119
+ ++ +Y+D FE+ FL+++ Y E QK ++ +Y+ V +R+ EEA+R
Sbjct 174 SLLSMLSDLQIYQDSFEQRFLEETNRLYAAEGQKLMQEREVPEYLHHVNKRLEEEADRLI 233
Query 120 HYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSG 179
YLD +T++ ++ VE +L+ H+ I++ G+ ++L+ +I+DL+ +Y+LF RV G
Sbjct 234 TYLDQTTQKSLIATVEKQLLGEHLTAILQ---KGLNNLLDENRIQDLSLLYQLFSRVRGG 290
Query 180 LKTII 184
++ ++
Sbjct 291 VQVLL 295
> At1g59790
Length=374
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 58/175 (33%), Positives = 101/175 (57%), Gaps = 2/175 (1%)
Query 6 VDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGI 65
+ S+ +GL F D V + ++ +E ++ L+ +ER GE IDR VKN + ++ G+
Sbjct 131 IPSLDEVGLTCFLDLV--YCEMQSTAKEVVIALIHKEREGEQIDRALVKNVLDIYVENGM 188
Query 66 NSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDAS 125
+ YE+DFE LQ +A +Y ++ ++ E ++ DY+ KVE+ + E ER HYL +
Sbjct 189 GTLEKYEEDFESFMLQDTASYYSRKASRWTEEDSCPDYMIKVEECLKMERERVTHYLHSI 248
Query 126 TEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGL 180
TE +V+ ++ EL+ K +E E+SG +L + K DL+R+Y+L+ + L
Sbjct 249 TEPKLVEKIQNELLVMVTKNRLENEHSGFSALLRDDKKNDLSRIYRLYLPIPKRL 303
> Hs4503161
Length=752
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 59/181 (32%), Positives = 96/181 (53%), Gaps = 15/181 (8%)
Query 9 VFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGIN-- 66
+++L L+ +RD + R + + +L+L+ +ERNGE I+ + ++LG+N
Sbjct 135 IYSLALVTWRDCLFR--PLNKQVTNAVLKLIEKERNGETINTRLISGVVQSYVELGLNED 192
Query 67 -------SRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC 119
+ TVY++ FE FL + FY ES +FL+ N +Y+KK E R+ EE R
Sbjct 193 DAFAKGPTLTVYKESFESQFLADTERFYTRESTEFLQQNPVTEYMKKAEARLLEEQRRVQ 252
Query 120 HYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSG 179
YL ST+ + + E LI H+ E+ ++ ++L+ K EDL RMY L R+ G
Sbjct 253 VYLHESTQDELARKCEQVLIEKHL----EIFHTEFQNLLDADKNEDLGRMYNLVSRIQDG 308
Query 180 L 180
L
Sbjct 309 L 309
> CE29089
Length=780
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 58/190 (30%), Positives = 104/190 (54%), Gaps = 20/190 (10%)
Query 5 NVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLG 64
N+ V+ L L++++ ++ F ++D + + +LEL+ ER G +I+ + L++LG
Sbjct 151 NIYMVYTLALVVWKRNL--FNDLKDKVIDAMLELIRSERTGSMINSRYISGVVECLVELG 208
Query 65 INSR-------------TVYEDDFEKHFLQQSAEFYKLESQKFLE-GNTACDYIKKVEQR 110
++ VY++ FE FL+ + FY E+ FL G DY+ KVE R
Sbjct 209 VDDSETDAKKDAETKKLAVYKEFFEVKFLEATRGFYTQEAANFLSNGGNVTDYMIKVETR 268
Query 111 INEEAERACHYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMY 170
+N+E +R YL++ST+ + E+ LISN + ++ G +L +++ +DL+RM+
Sbjct 269 LNQEDDRCQLYLNSSTKTPLATCCESVLISNQLD-FLQRHFGG---LLVDKRDDDLSRMF 324
Query 171 KLFGRVDSGL 180
KL RV +GL
Sbjct 325 KLCDRVPNGL 334
> 7304137
Length=774
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 53/188 (28%), Positives = 93/188 (49%), Gaps = 25/188 (13%)
Query 3 QNNVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQ 62
Q + ++ L L+ ++ H+ F ++ + + + +L+ + ER G+LI+R V++ ++
Sbjct 144 QKGIYKIYRLALVAWKGHL--FQVLNEPVTKAVLKSIEEERQGKLINRSLVRDVIECYVE 201
Query 63 LGINSR---------TVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINE 113
L N +VY+ +FE F+ ++ FY+ ES FL NT +Y+K VE R+ E
Sbjct 202 LSFNEEDTDAEQQKLSVYKQNFENKFIADTSAFYEKESDAFLSTNTVTEYLKHVENRLEE 261
Query 114 EAER--------ACHYLDASTEQLIVKVVETELISNHMKTI-VEMENSGVVHMLENQKIE 164
E +R YL +T ++ E LI H+K E +N +L + +
Sbjct 262 ETQRVRGFNSKNGLSYLHETTADVLKSTCEEVLIEKHLKIFHTEFQN-----LLNADRND 316
Query 165 DLARMYKL 172
DL RMY L
Sbjct 317 DLKRMYSL 324
> At1g59800
Length=255
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 43/131 (32%), Positives = 74/131 (56%), Gaps = 2/131 (1%)
Query 3 QNNVDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQ 62
+ + S+ +GL FRD V R ++ E +L L+ +ER GE IDR V+N + ++
Sbjct 119 KKGLPSLREVGLNCFRDQVYR--EMQSMAAEAILALIHKEREGEQIDRELVRNVIDVFVE 176
Query 63 LGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYL 122
G+ + YE+DFE+ LQ +A +Y ++ ++++ + DY K +Q + E ER HYL
Sbjct 177 NGMGTLKKYEEDFERLMLQDTASYYSSKASRWIQEESCLDYTLKPQQCLQRERERVTHYL 236
Query 123 DASTEQLIVKV 133
+TE + +V
Sbjct 237 HPTTEPKLFEV 247
> SPAC17G6.12
Length=767
Score = 75.5 bits (184), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 54/185 (29%), Positives = 92/185 (49%), Gaps = 13/185 (7%)
Query 6 VDSVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGI 65
V ++ L L+ + HV F IRD + + LL + ++R E D V+ + L
Sbjct 151 VYDIYTLCLVSWHHHV--FSHIRDSLLQNLLYMFTKKRLYEPTDMKYVEVCVDSITSLSF 208
Query 66 NSR-------TVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERA 118
+ + Y+ FE +F++ + FY ES ++L ++ DY+KK E R+ EE E
Sbjct 209 DKTDMTKPNLSSYKTFFETNFIENTKNFYAKESSEYLASHSITDYLKKAEIRLAEEEELV 268
Query 119 CHYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDS 178
YL ST + +++ E LI+ H E+ ++ ML+ ED+ RMY+L R +
Sbjct 269 RLYLHESTLKPLLEATEDVLIAQHE----EVLHNDFARMLDQNCSEDIIRMYRLMSRTPN 324
Query 179 GLKTI 183
GL+ +
Sbjct 325 GLQPL 329
> SPAC24H6.03
Length=785
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 56/174 (32%), Positives = 96/174 (55%), Gaps = 3/174 (1%)
Query 13 GLILFRDHVVRFGL-IRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSR--T 69
G+ +FR+ V+ I + ET+L LV ER G I+R + + ML L ++ T
Sbjct 163 GIYIFREVVLLNSFEIGEKCVETILILVYLERKGNTINRPLINDCLDMLNSLPSENKKET 222
Query 70 VYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQL 129
+Y+ F FL + FY++ES + +Y+KK E+R EE ER+ +YL
Sbjct 223 LYDVLFAPKFLSYTRNFYEIESSTVIGVFGVVEYLKKAEKRFEEEKERSKNYLFTKIASP 282
Query 130 IVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTI 183
++ VVE EL+S H+ ++E +++G M+++ E L +Y+ F RV+ G+K++
Sbjct 283 LLSVVEDELLSKHLDDLLENQSTGFFSMIDSSNFEGLQLVYESFSRVELGVKSL 336
> CE11928
Length=729
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 51/148 (34%), Positives = 75/148 (50%), Gaps = 11/148 (7%)
Query 33 ETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQ 92
+ +LEL++ ER G I+ + + L +L I Y+ FE FL + FYK E
Sbjct 152 DAILELILLERCGSTINSTHISSVVECLTELDI-----YKVSFEPQFLDATKLFYKQEVL 206
Query 93 KFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQLIVKVVETELISNHMKTIVEMENS 152
E T +Y+ VE R+ +E R+ YL ST L++ E+ LIS+ +K + +S
Sbjct 207 NSKE--TVIEYMITVENRLFQEEYRSRRYLGPSTNDLLIDSCESILISDRLKFL----HS 260
Query 153 GVVHMLENQKIEDLARMYKLFGRVDSGL 180
+LE +K E L RMY L RV GL
Sbjct 261 EFERLLEARKDEHLTRMYSLCRRVTHGL 288
> Hs4503167
Length=780
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 43/168 (25%), Positives = 87/168 (51%), Gaps = 11/168 (6%)
Query 24 FGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSR---TVYEDDFEKHFL 80
F I++ ++++ ++LV ER GE D V + L N +Y D+FEK +L
Sbjct 148 FSNIKNRLQDSAMKLVHAERLGEAFDSQLVIGVRESYVNLCSNPEDKLQIYRDNFEKAYL 207
Query 81 QQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTE----QLIVKVVET 136
+ FY+ ++ +L+ N +Y+K + ++ EE +RA YL+ E + +++
Sbjct 208 DSTERFYRTQAPSYLQPNGVQNYMKYADAKLKEEEKRALRYLETRRECNSVEALMECCVN 267
Query 137 ELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTII 184
L+++ +TI+ E G M++ + E L M+ L +V +G++ ++
Sbjct 268 ALVTSFKETIL-AECQG---MIKRNETEKLHLMFSLMDKVPNGIEPML 311
> 7301740
Length=852
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 42/160 (26%), Positives = 83/160 (51%), Gaps = 7/160 (4%)
Query 24 FGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSRT---VYEDDFEKHFL 80
F I+ ++E+ +++V ERNG+ D V + L N+ +Y ++FE +L
Sbjct 226 FHDIKHRLQESAMKIVHAERNGDAYDAQLVVGVRESYVNLSSNAEDKLEIYRENFEMAYL 285
Query 81 QQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQLIVKVVETELIS 140
+ + EFY+L+S + + N Y+K + ++ EE RA YL+ S+ ++ + LI
Sbjct 286 KATVEFYRLKSAEQQQENGVLAYMKYADSKLREEEVRAKRYLEPSSFSILTYTLVNVLIV 345
Query 141 NHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGL 180
+H+ +I+ + ++ + + E L M++L RV G+
Sbjct 346 DHLNSII----AECPALIRDYETERLNLMFRLMDRVMHGV 381
> YDL132w
Length=815
Score = 62.4 bits (150), Expect = 6e-10, Method: Composition-based stats.
Identities = 45/161 (27%), Positives = 81/161 (50%), Gaps = 11/161 (6%)
Query 22 VRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSR-------TVYEDD 74
V F +D + LL+ V R G++I R + L+ LGI+ + VY
Sbjct 164 VMFDPSKDVLINELLDQVTLGREGQIIQRSNISTAIKSLVALGIDPQDLKKLNLNVYIQV 223
Query 75 FEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQLIVKVV 134
FEK FL+++ E+Y + +LE ++ +YI + + I E + Y D T++ + +
Sbjct 224 FEKPFLKKTQEYYTQYTNDYLEKHSVTEYIFEAHEIIKREEKAMTIYWDDHTKKPLSMAL 283
Query 135 ETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGR 175
LI++H++ ++EN VV +L+ + IE + +Y L R
Sbjct 284 NKVLITDHIE---KLENEFVV-LLDARDIEKITSLYALIRR 320
> CE06658
Length=741
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/165 (26%), Positives = 81/165 (49%), Gaps = 20/165 (12%)
Query 36 LELVIRERNGELIDRLAVKNTCHMLIQL----GINSRTVYEDDFEKHFLQQSAEFYKLES 91
L LV ER+G +ID V + L G + VY FE+ F++Q+ E+YK
Sbjct 144 LRLVKEERDGNIIDAQNVIGIRESFVALNDRAGEDPLLVYRQSFERQFIEQTTEYYKKIC 203
Query 92 QKFLEGNTACDYIKKVEQRINEEAERACHYLDAST-------EQLIVKVVETELISNHMK 144
L +Y+ ++++ EE +RA YL+ ++ E+ ++ +VE+
Sbjct 204 GNLLNELGVLEYMVYADKKLEEEQQRAKRYLEMNSPTSGKHMEKAVIALVES-----FED 258
Query 145 TIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTIIACVST 189
TI+ + ++ ++ +E L R+Y+L R SG+ T++ C+ T
Sbjct 259 TIL----AECSKLIASKDVERLQRLYRLIRRTRSGIDTVLKCIDT 299
> At4g12100
Length=434
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 47/180 (26%), Positives = 92/180 (51%), Gaps = 23/180 (12%)
Query 8 SVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHML------I 61
+++++G LF + ++D + +L L+ ER G+ A NT +L
Sbjct 190 TLWDVGQKLFHKQLSMAPQLQDQVITGILRLITDERLGK-----AANNTSDLLKNLMDMF 244
Query 62 QLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC-- 119
++ VY+D F L +++FY E+++ L+ + Y+K VE+ E E+ C
Sbjct 245 RMQWQCTYVYKDPF----LDSTSKFYAEEAEQVLQRSDISHYLKYVERTFLAEEEK-CDK 299
Query 120 -HYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDS 178
++ +S+ ++KV++++L+ H + E G + +++ I+DL RMY+LF VDS
Sbjct 300 HYFFFSSSRSRLMKVLKSQLLEAHSSFLEE----GFMLLMDESLIDDLRRMYRLFSMVDS 355
> At4g02570
Length=676
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 50/76 (65%), Gaps = 0/76 (0%)
Query 108 EQRINEEAERACHYLDASTEQLIVKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLA 167
E+ + +E ER HYL +S+E +V+ V+ EL+ ++E E+SG +L + K++DL+
Sbjct 163 EECLKKERERVAHYLHSSSEPKLVEKVQHELLVVFASQLLEKEHSGCRALLRDDKVDDLS 222
Query 168 RMYKLFGRVDSGLKTI 183
RMY+L+ ++ GL+ +
Sbjct 223 RMYRLYHKILRGLEPV 238
> Hs19482174
Length=745
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 62/114 (54%), Gaps = 4/114 (3%)
Query 71 YEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQLI 130
Y++ FE FL ++ E+YK E+ L+ + Y++KV R+ +E R YL S+ +
Sbjct 203 YQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKVLGRLKDEEIRCRKYLHPSSYTKV 262
Query 131 VKVVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTII 184
+ + ++++H++ + ++ +++ +K D+A MY L V +GL +I
Sbjct 263 IHECQQRMVADHLQFL----HAECHNIIRQEKKNDMANMYVLLRAVSTGLPHMI 312
> At1g02980
Length=648
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 44/160 (27%), Positives = 72/160 (45%), Gaps = 22/160 (13%)
Query 25 GLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGINSRTVYEDDFEKHFLQQSA 84
G+ D+ ++T+L IRE++GE + R VK + I + S HF +
Sbjct 69 GVYVDYNKQTVLP-AIREKHGEYMLRELVKRWANQKILVRWLS----------HFFEYLD 117
Query 85 EFY-KLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQLIVKVVETELISNHM 143
FY + S L + V Q + +A+ A L E L+V
Sbjct 118 RFYTRRGSHPTLSAVGFISFRDLVYQELQSKAKDAVLALKVQNELLVVVA---------- 167
Query 144 KTIVEMENSGVVHMLENQKIEDLARMYKLFGRVDSGLKTI 183
K ++E E+SG +L + K++DLARMY+L+ + GL +
Sbjct 168 KQLIENEHSGCRALLRDDKMDDLARMYRLYHPIPQGLDPV 207
> CE16729
Length=745
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 62/120 (51%), Gaps = 6/120 (5%)
Query 66 NSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDAS 125
++ Y++ FEK L + ++Y +QK L + +Y+++V + +E RA YL S
Sbjct 231 STTAFYQESFEKPLLTDTEQYYSALAQKMLTDLSCSEYMEQVIVLLEQEEMRAKKYLHES 290
Query 126 TEQLIVKVVETELISNHMKTIVEMENSGVVH-MLENQKIEDLARMYKLFGRVDSGLKTII 184
+ + ++ + + +I H + V H ++ N++ +DL MY+L + +GL ++
Sbjct 291 SVEKVITLCQKVMIKAHKDKL-----HAVCHDLITNEENKDLRNMYRLLKPIQAGLSVMV 345
> CE24730
Length=728
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 62/120 (51%), Gaps = 6/120 (5%)
Query 66 NSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDAS 125
++ Y++ FEK L + ++Y +QK L + +Y+++V + +E RA YL S
Sbjct 214 STTAFYQESFEKPLLTDTEQYYSALAQKMLTDLSCSEYMEQVIVLLEQEEMRAKKYLHES 273
Query 126 TEQLIVKVVETELISNHMKTIVEMENSGVVH-MLENQKIEDLARMYKLFGRVDSGLKTII 184
+ + ++ + + +I H + V H ++ N++ +DL MY+L + +GL ++
Sbjct 274 SVEKVITLCQKVMIKAHKDKL-----HAVCHDLITNEENKDLRNMYRLLKPIQAGLSVMV 328
> CE25691
Length=804
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 62/120 (51%), Gaps = 6/120 (5%)
Query 66 NSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDAS 125
++ Y++ FEK L + ++Y +QK L + +Y+++V + +E RA YL S
Sbjct 290 STTAFYQESFEKPLLTDTEQYYSALAQKMLTDLSCSEYMEQVIVLLEQEEMRAKKYLHES 349
Query 126 TEQLIVKVVETELISNHMKTIVEMENSGVVH-MLENQKIEDLARMYKLFGRVDSGLKTII 184
+ + ++ + + +I H + V H ++ N++ +DL MY+L + +GL ++
Sbjct 350 SVEKVITLCQKVMIKAHKDKL-----HAVCHDLITNEENKDLRNMYRLLKPIQAGLSVMV 404
> At3g46910
Length=247
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 57/112 (50%), Gaps = 11/112 (9%)
Query 8 SVFNLGLILFRDHVVRFGLIRDHIRETLLELVIRERNGELIDRLAVKNTCHMLIQLGIN- 66
SV++LG L H+ +RD + +L+L+ +R+ +D +KNT ++ + +
Sbjct 133 SVWDLGSELSPKHLFSAQKVRDKLLSIILQLIRDQRSFMSVDMTQLKNTTRPVMSVHMTQ 192
Query 67 ---------SRTVYEDDF-EKHFLQQSAEFYKLESQKFLEGNTACDYIKKVE 108
+++Y+ F +K F+ + EFY E+ +F E + Y+K+VE
Sbjct 193 LNNLRGLFYGQSLYKSPFFKKPFIDCAVEFYSAEAMQFKEQSDIPLYLKRVE 244
> HsM13376093
Length=825
Score = 32.3 bits (72), Expect = 0.57, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Query 55 NTCHMLIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFL 95
+ CHML+Q G N T ED ++ L ++AE LE+ K+L
Sbjct 756 DICHMLVQAGANIDTCSED--QRTPLMEAAENNHLEAVKYL 794
> Hs21314722
Length=1267
Score = 32.3 bits (72), Expect = 0.57, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Query 55 NTCHMLIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFL 95
+ CHML+Q G N T ED ++ L ++AE LE+ K+L
Sbjct 756 DICHMLVQAGANIDTCSED--QRTPLMEAAENNHLEAVKYL 794
> Hs4504951
Length=1786
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 21/99 (21%), Positives = 47/99 (47%), Gaps = 6/99 (6%)
Query 73 DDFEKHFLQQSAEFYKLESQKFLEGNTACDYIKKVEQRINEEAERACHYLDASTEQLIVK 132
++ ++ Q S E +E + +A D K ++ ++E+ ++ + + TE+
Sbjct 1656 EELKRKAAQNSGEAEYIEKVVYTVKQSAEDVKKTLDGELDEKYKKVENLIAKKTEESADA 1715
Query 133 VVETELISNHMKTIVEMENSGVVHMLENQKIEDLARMYK 171
+ E++ N KT++ NS + Q ++DL R Y+
Sbjct 1716 RRKAEMLQNEAKTLLAQANSKL------QLLKDLERKYE 1748
> At3g50400
Length=374
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 44/96 (45%), Gaps = 11/96 (11%)
Query 28 RDHIRE-TLLELVIRER---NGELIDRLAVKNTCHMLIQLGINSRTVYEDDFEKHFLQQS 83
RD+IR+ +L +VI N L+ +A + L Q + + DD H Q
Sbjct 161 RDYIRKRSLFSVVIGSNDFLNNYLVPFVAAQ---ARLTQ----TPETFVDDMISHLRNQL 213
Query 84 AEFYKLESQKFLEGNTACDYIKKVEQRINEEAERAC 119
Y ++++KF+ GN A ++ IN+ ++ C
Sbjct 214 KRLYDMDARKFVVGNVAPIGCIPYQKSINQLNDKQC 249
> At2g35060
Length=792
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 41/148 (27%), Positives = 66/148 (44%), Gaps = 26/148 (17%)
Query 47 LIDRLAVKNTCHM---LIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEG--NTAC 101
L+ R+ KN HM + + G +DDFEK + + +LES +EG + +
Sbjct 610 LVKRIGPKNF-HMFRCVARYGYRDLHKKDDDFEKRLFESLFLYVRLES--MMEGGCSDSD 666
Query 102 DY-IKKVEQRI-------NEEAERACHYLDASTEQL--IVKVVETELISNHMKTIVEME- 150
DY I +Q++ NE A S E + + +V T S+ M + E+E
Sbjct 667 DYSICGSQQQLKDTLGNGNENENLATFDTFDSIESITPVKRVSNTVTASSQMSGVDELEF 726
Query 151 -----NSGVVHMLENQKIEDL--ARMYK 171
++GVVH++ N + AR YK
Sbjct 727 INGCRDAGVVHIMGNTVVRARREARFYK 754
> Hs20537850
Length=710
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 41/96 (42%), Gaps = 23/96 (23%)
Query 58 HMLIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGNTACD--------------- 102
H + + S Y++ K LQ+ A F +L +Q LE + C
Sbjct 359 HFSVYITYVSNQTYQERTYKQLLQEKAAFRELIAQ--LELDPKCRGLPFSSFLILPFQRI 416
Query 103 -----YIKKVEQRINEEAERACHYLDASTE-QLIVK 132
++ + +R+ E +ER C LDA E +++VK
Sbjct 417 TRLKLLVQNILKRVEERSERECTALDAHKELEMVVK 452
> CE09154
Length=542
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 37/74 (50%), Gaps = 0/74 (0%)
Query 39 VIRERNGELIDRLAVKNTCHMLIQLGINSRTVYEDDFEKHFLQQSAEFYKLESQKFLEGN 98
V +ER E DR+ ++LI G N+++++ D K L ++ E + + N
Sbjct 288 VKQERLPEEYDRVLSLRKKNVLISFGTNAKSMFMSDDMKESLIKTFESMPDTTFIWKYEN 347
Query 99 TACDYIKKVEQRIN 112
T D +K+ +RIN
Sbjct 348 TTVDIVKQYNKRIN 361
Lambda K H
0.322 0.138 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3130490202
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40