bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0051_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
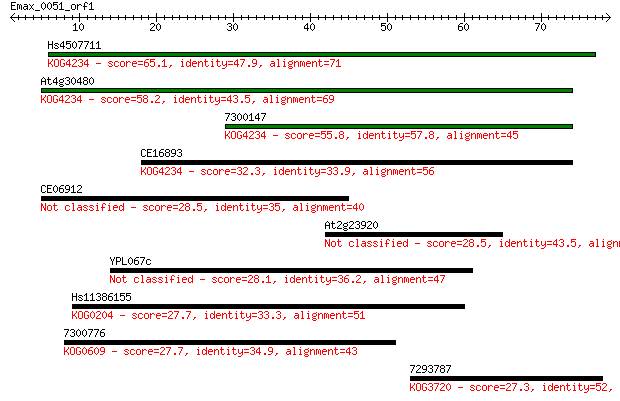
Hs4507711 65.1 3e-11
At4g30480 58.2 3e-09
7300147 55.8 2e-08
CE16893 32.3 0.19
CE06912 28.5 3.4
At2g23920 28.5 3.4
YPL067c 28.1 4.4
Hs11386155 27.7 4.7
7300776 27.7 5.1
7293787 27.3 7.2
> Hs4507711
Length=292
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Query 6 DPKLSSTYGGELQRVKRESEALFEKEKEEMMGKLKDLGNFVLGKVGLSLDNFKVEQNPET 65
DP + R+ ++ E E+ KEEM+GKLKDLGN VL GLS +NF+++Q+ T
Sbjct 219 DPSIHQAREA-CMRLPKQIEERNERLKEEMLGKLKDLGNLVLRPFGLSTENFQIKQDSST 277
Query 66 GSYNIQFQQNP 76
GSY+I F QNP
Sbjct 278 GSYSINFVQNP 288
> At4g30480
Length=277
Score = 58.2 bits (139), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 5 LDPKLSSTYGGELQRVKRESEALFEKEKEEMMGKLKDLGNFVLGKVGLSLDNFKVEQNPE 64
LDP G ++R++ + EK KEE + KLK++GN +LG+ G+S+DNFK ++P
Sbjct 209 LDPSNDQARKG-IRRLEPLAAEKREKMKEEAITKLKEMGNSILGRFGMSVDNFKAVKDPN 267
Query 65 TGSYNIQFQ 73
TGSY++ FQ
Sbjct 268 TGSYSLSFQ 276
> 7300147
Length=256
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/45 (57%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 29 EKEKEEMMGKLKDLGNFVLGKVGLSLDNFKVEQNPETGSYNIQFQ 73
EK K EMM LKDLGN +L GLS NF+++Q+P TGSY+I F+
Sbjct 212 EKLKNEMMSSLKDLGNMILKPFGLSTQNFQMQQDPNTGSYSINFK 256
> CE16893
Length=207
Score = 32.3 bits (72), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 5/56 (8%)
Query 18 QRVKRESEALFEKEKEEMMGKLKDLGNFVLGKVGLSLDNFKVEQNPETGSYNIQFQ 73
+++ +EA+ K ++M KLK GNF L GLS D+F++ N G +++Q +
Sbjct 138 EKINNRNEAM----KADVMEKLKGFGNFCLSPFGLSTDSFEMVPNG-NGGFSVQMK 188
> CE06912
Length=142
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 5 LDPKLSSTYGGELQRVKRESEALFEKEKEEMMGKLKDLGN 44
L+ ++ G EL RVKR S+ EK+ +E+M K++ + +
Sbjct 27 LESDVAEMEGAELDRVKRASDHKLEKKLDEIMEKVEKIKD 66
> At2g23920
Length=56
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 10/23 (43%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 42 LGNFVLGKVGLSLDNFKVEQNPE 64
+GN + G+ G+S+DNFK + P
Sbjct 1 MGNSIFGRCGMSVDNFKAVKIPT 23
> YPL067c
Length=198
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 4/49 (8%)
Query 14 GGELQRVKRESEAL--FEKEKEEMMGKLKDLGNFVLGKVGLSLDNFKVE 60
GEL R+KR + + + K+ G D+ +VL K+G SLD ++E
Sbjct 27 SGELARLKRSRQMTDKYHEHKKRTAGL--DMNQYVLQKLGWSLDEPQLE 73
> Hs11386155
Length=1173
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 9 LSSTYGGELQRVKRESEALFEKEKEEMMGKLKDLGNFVLGKVGLSLDNFKV 59
L S GGE++ +++ +KEK + GKL L +GK GL + V
Sbjct 334 LKSAEGGEMEEREKKKANAPKKEKSVLQGKLTKLA-VQIGKAGLVMSAITV 383
> 7300776
Length=461
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query 8 KLSSTYGGELQRVKRESEALFEKEKEEMMG---KLKDLGNFVLGKV 50
+L + GGELQ V R F+K +E MG K+ + G ++ ++
Sbjct 41 ELDNVEGGELQHVTRVRLVQFQKNTDEPMGITLKMTEDGRCIVARI 86
> 7293787
Length=410
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Query 53 SLDNFKVEQNPETGSY--NIQFQQNPK 77
SL F++ QNP+TG Y I FQ +P
Sbjct 333 SLIAFELHQNPQTGEYFLEIYFQNDPH 359
Lambda K H
0.311 0.134 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171925608
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40