bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0063_orf2
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
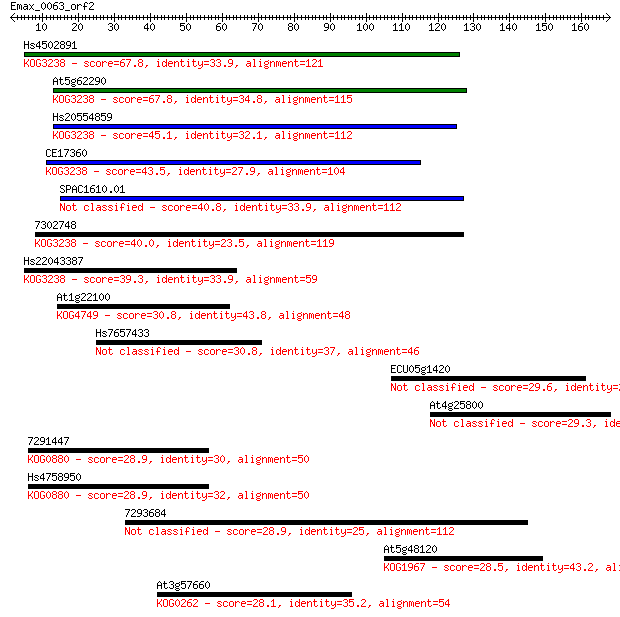
Hs4502891 67.8 9e-12
At5g62290 67.8 9e-12
Hs20554859 45.1 7e-05
CE17360 43.5 2e-04
SPAC1610.01 40.8 0.001
7302748 40.0 0.002
Hs22043387 39.3 0.004
At1g22100 30.8 1.2
Hs7657433 30.8 1.2
ECU05g1420 29.6 3.0
At4g25800 29.3 4.2
7291447 28.9 4.5
Hs4758950 28.9 5.2
7293684 28.9 5.5
At5g48120 28.5 7.2
At3g57660 28.1 7.8
> Hs4502891
Length=237
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 58/121 (47%), Gaps = 10/121 (8%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEA 64
++GK G G L+I R++WL G F+L Y +I LHALS D C LY + +
Sbjct 27 LNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPTISLHALSRDRSDCLGEHLYVMVNAK- 85
Query 65 LADLSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
S + EE + D + + E +FVP D S L+ +FT M + ALH
Sbjct 86 ---------FEEESKEPVADEEEEDSDDDVEPITEFRFVPSDKSALEAMFTAMCECQALH 136
Query 125 P 125
P
Sbjct 137 P 137
> At5g62290
Length=229
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 62/117 (52%), Gaps = 16/117 (13%)
Query 13 GELFITSQRVAWLSDV--GASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEALADLSN 70
G L+ITS+++ WLSDV +A++++SI LHA+S DP+A PC+Y QI
Sbjct 49 GTLYITSRKLIWLSDVDMAKGYAVDFLSISLHAVSRDPEAYSSPCIYTQI---------- 98
Query 71 GNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALHPDP 127
+ E+ E D + E++ VP D + L+ LF V + A L+P+P
Sbjct 99 ----EVEEDEDDESDSESTEVLDLSKIREMRLVPSDSTQLETLFDVFCECAELNPEP 151
> Hs20554859
Length=220
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 49/112 (43%), Gaps = 12/112 (10%)
Query 13 GELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEALADLSNGN 72
G L+I ++WL G F+L Y +I L ALS D C LY + +
Sbjct 82 GTLYIAESHLSWLDSSGLGFSLEYPTISLLALSRDQSDCLGEHLYATVNDKF-------- 133
Query 73 PVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALH 124
S + EE E+ D + + E FVP D S L +FT M + ALH
Sbjct 134 ----EESKESVADEEEEDSDDVELITEFIFVPSDKSALGAMFTAMCECQALH 181
> CE17360
Length=205
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 14/107 (13%)
Query 11 GRGELFITSQRVAWLSDVGAS--FALNYVSIVLHALSTDPQACDRPCLYCQI-KGEALAD 67
G G L+IT V W+S + F++ Y +IVLHA+STD ++ + + ++ +
Sbjct 30 GNGTLYITDSAVIWISSAAGTKGFSVAYPAIVLHAISTDVSVFPSEHIFVMVDQRKSGLE 89
Query 68 LSNGNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLF 114
L+ S D P +E++FVPDD L +++
Sbjct 90 LAAAELEDEESDDDEEEP-----------ALEIRFVPDDKDSLSQIY 125
> SPAC1610.01
Length=217
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 57/116 (49%), Gaps = 13/116 (11%)
Query 15 LFITSQR-VAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGEALADLSN--- 70
L +TSQ + + + + + + Y +I LHA Q+ D+P +Y Q++GEA+ L +
Sbjct 55 LLVTSQSFILFDEEQNSGWKIPYETITLHA----KQSKDKPYVYVQLEGEAIRPLLDHIL 110
Query 71 GNPVSSSSSDSACMPEEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALHPD 126
SS + A E+ E DD + L +V D SC Q L T S LHPD
Sbjct 111 KFERSSGTLHEAPSTEDENEFTDDFLELTL-YVTDVDSCYQALCTCQS----LHPD 161
> 7302748
Length=215
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/135 (20%), Positives = 57/135 (42%), Gaps = 16/135 (11%)
Query 8 KDAGRGELFITSQRVAWL-SDVGASFALNYVSIVLHALSTDPQAC-----DRPCLYCQIK 61
K G G ++I ++W +++ ++ + + LH +S++P+ C D + +
Sbjct 28 KVVGEGTVYIAQNTLSWQPTELAEGISIEWKQVSLHGISSNPRKCIYFMLDHKVEWNGVY 87
Query 62 GEALADLSNGNPVSSSS--------SDSACMPEEAEECADDD--AMVELKFVPDDPSCLQ 111
G+ NG S SD + E+ D+ + E +P+D +
Sbjct 88 GDPPQQAVNGRNGGGSEAEVDEGNGSDEHDEDDNFEDAVDEQFGEVTECWLMPEDIHTVD 147
Query 112 RLFTVMSDMAALHPD 126
+++ M+ ALHPD
Sbjct 148 TMYSAMTTCQALHPD 162
> Hs22043387
Length=235
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 5 ISGKDAGRGELFITSQRVAWLSDVGASFALNYVSIVLHALSTDPQACDRPCLYCQIKGE 63
++GK G L+I ++WL G F+L +I LHALS D LY +K +
Sbjct 35 VNGKGLSTGTLYIAESHLSWLDGSGLRFSLECPTICLHALSRDQSDYLGEHLYVMVKAK 93
> At1g22100
Length=457
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 27/51 (52%), Gaps = 4/51 (7%)
Query 14 ELFITSQRVAWLSDVGASFALNYVSIVLH---ALSTDPQACDRPCLYCQIK 61
E ITSQR +W +DV AS N S++L L D+PCL +IK
Sbjct 134 EKIITSQRPSWRADV-ASVDTNRSSVLLMDDLTLFAHGHVEDKPCLSVEIK 183
> Hs7657433
Length=880
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query 25 LSDVGASFALNYVSIVLHAL-STDPQACDRPCLYCQIKGEALADLSN 70
LS G S Y +++LHA +T CD P L+C+++ + LA+L +
Sbjct 182 LSTPGHSPHSAYTTLLLHAFQATFGAHCDVPGLHCRLQAKTLAELED 228
> ECU05g1420
Length=319
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 107 PSCLQRLFTVMSDMAALHPDPDSATLDGDEDELFDEASLRRQGWEFVENEDDMG 160
P C+ R+ ++S + L T + DE+ + S R +E N+D MG
Sbjct 31 PLCIGRINLILSKVVKLRVQKVRRTESDNNDEIEETCSGYRHKFEVYSNDDPMG 84
> At4g25800
Length=536
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 26/56 (46%), Gaps = 8/56 (14%)
Query 118 SDMAALHPDPDSATLDGDEDELFDEASLRRQGWEFVENED------DMGGSGGMDG 167
+D L+P D L E++ F E +R+ + + NED MGG G DG
Sbjct 418 ADQENLNPFEDWTNLS--ENDFFSEEEIRQTSHDLLANEDMQQLLFSMGGGKGEDG 471
> 7291447
Length=205
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 6 SGKDAGRGELFITSQRVAWLSDVGASFA--LNYVSIVLHALSTDPQACDRPC 55
+GKD + FIT+++ +WL F L+ +++V ++ A DRP
Sbjct 128 AGKDTNGSQFFITTKQTSWLDGRHVVFGKILSGMNVVRQIENSATDARDRPV 179
> Hs4758950
Length=216
Score = 28.9 bits (63), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 24/52 (46%), Gaps = 2/52 (3%)
Query 6 SGKDAGRGELFITSQRVAWLSDVGASF--ALNYVSIVLHALSTDPQACDRPC 55
+GKD + FIT+ + AWL F L + +V ST + D+P
Sbjct 143 AGKDTNGSQFFITTVKTAWLDGKHVVFGKVLEGMEVVRKVESTKTDSRDKPL 194
> 7293684
Length=350
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 44/119 (36%), Gaps = 9/119 (7%)
Query 33 ALNYVSIVLHALSTDP-------QACDRPCLYCQIKGEALADLSNGNPVSSSSSDSACMP 85
Y SIV + ++ D C++ L + + S G VS + D P
Sbjct 162 TFEYCSIVKNGVNFDNLQGCNMYHVCEKGVLKDKTCSKTYYQASTGECVSKALVDCDAHP 221
Query 86 EEAEECADDDAMVELKFVPDDPSCLQRLFTVMSDMAALHPDPDSATLDGDEDELFDEAS 144
+ C E KFV D+ +C R + + PDP+ +D FD +S
Sbjct 222 LPTDVCGKASKPYENKFVADEATC--RGYFYCAKQKDGTPDPNPVWNQCPQDRFFDASS 278
> At5g48120
Length=1152
Score = 28.5 bits (62), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 27/60 (45%), Gaps = 17/60 (28%)
Query 105 DDPSCLQRLFTVMSDMAALHPDPDSATLDGDEDELF----------------DEASLRRQ 148
DP CL +F ++ +A L P P S L D +LF DEA++RR+
Sbjct 189 KDPQCLMIVFHLVELLAPLFPSP-SGPLASDASDLFEVIGCYFPLHFTHTKDDEANIRRE 247
> At3g57660
Length=1670
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 28/62 (45%), Gaps = 8/62 (12%)
Query 42 HALSTDPQACDRPC--LYCQIKGEALA--DLSNGNPVSSSSSDSAC----MPEEAEECAD 93
H P+ +R L IKG+ ++ L + P S SSD +C + +EEC D
Sbjct 123 HHFMAKPEDVERAVSQLKLIIKGDIVSAKQLESNTPTKSKSSDESCESVVTTDSSEECED 182
Query 94 DD 95
D
Sbjct 183 SD 184
Lambda K H
0.315 0.132 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2425968562
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40