bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
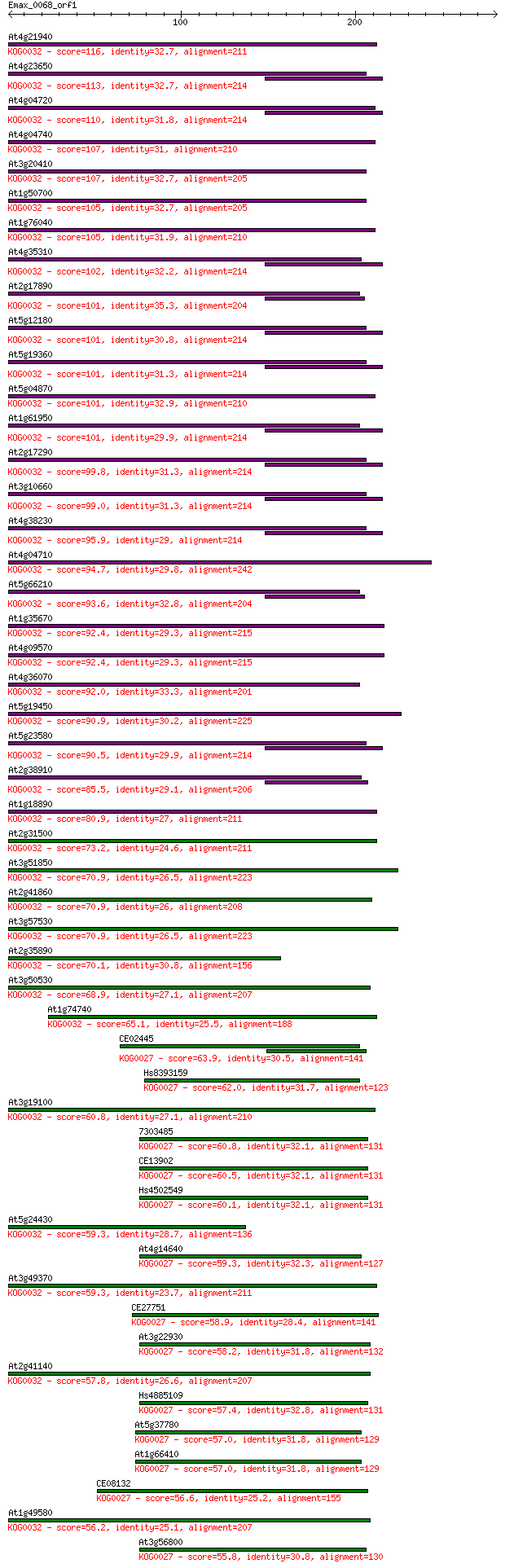
Query= Emax_0068_orf1
Length=280
Score E
Sequences producing significant alignments: (Bits) Value
At4g21940 116 6e-26
At4g23650 113 5e-25
At4g04720 110 4e-24
At4g04740 107 3e-23
At3g20410 107 3e-23
At1g50700 105 8e-23
At1g76040 105 8e-23
At4g35310 102 1e-21
At2g17890 101 1e-21
At5g12180 101 2e-21
At5g19360 101 2e-21
At5g04870 101 2e-21
At1g61950 101 2e-21
At2g17290 99.8 6e-21
At3g10660 99.0 1e-20
At4g38230 95.9 8e-20
At4g04710 94.7 2e-19
At5g66210 93.6 4e-19
At1g35670 92.4 8e-19
At4g09570 92.4 1e-18
At4g36070 92.0 1e-18
At5g19450 90.9 2e-18
At5g23580 90.5 4e-18
At2g38910 85.5 1e-16
At1g18890 80.9 3e-15
At2g31500 73.2 5e-13
At3g51850 70.9 3e-12
At2g41860 70.9 3e-12
At3g57530 70.9 3e-12
At2g35890 70.1 5e-12
At3g50530 68.9 1e-11
At1g74740 65.1 1e-10
CE02445 63.9 3e-10
Hs8393159 62.0 1e-09
At3g19100 60.8 3e-09
7303485 60.8 3e-09
CE13902 60.5 3e-09
Hs4502549 60.1 5e-09
At5g24430 59.3 8e-09
At4g14640 59.3 9e-09
At3g49370 59.3 9e-09
CE27751 58.9 1e-08
At3g22930 58.2 2e-08
At2g41140 57.8 3e-08
Hs4885109 57.4 4e-08
At5g37780 57.0 4e-08
At1g66410 57.0 4e-08
CE08132 56.6 6e-08
At1g49580 56.2 8e-08
At3g56800 55.8 9e-08
> At4g21940
Length=554
Score = 116 bits (290), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 69/214 (32%), Positives = 116/214 (54%), Gaps = 5/214 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + LL ++P+ R++AA ALEHPW + G ID VL M++F A +++
Sbjct 333 AKDLVRKLLTKDPKQRISAAQALEHPW--IRGGEAPDKPIDSAVLSRMKQFRAMNKLKKL 390
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA S+ +++ L+ +F +D K+G+I E L N L K + + EAE ++
Sbjct 391 ALKVIAESLSEEEIKGLKTMFANMDTDKSGTITYEELKNGLAKLGSKLTEAEVKQLMEAA 450
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGGV 179
D+ G+ I+Y EF++AT + +AF+ D D SG I++D L + + G
Sbjct 451 DVDGNGTIDYIEFISATMHRYRFDRDEHVFKAFQYFDKDNSGFITMDELESAMKEYGMGD 510
Query 180 LA--EEILQQCDIKKNGVIDFEEFMVALTGAVTF 211
A +E++ + D +G I++EEF + +T
Sbjct 511 EASIKEVIAEVDTDNDGRINYEEFCAMMRSGITL 544
> At4g23650
Length=529
Score = 113 bits (282), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 68/211 (32%), Positives = 117/211 (55%), Gaps = 10/211 (4%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L+ +P+DRLTAA L HPW+ EDG +D VL M++F A +++
Sbjct 309 AKDLVRKMLKYDPKDRLTAAEVLNHPWIR-EDGEASDKPLDNAVLSRMKQFRAMNKLKKM 367
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA ++ +++ L+++F +LD G + +E L L K + I EAE ++
Sbjct 368 ALKVIAENLSEEEIIGLKEMFKSLDTDNNGIVTLEELRTGLPKLGSKISEAEIRQLMEAA 427
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNL-----RHILGD 174
D+ GD I+Y EF++AT + + AF+ D D SG I+++ L ++ +GD
Sbjct 428 DMDGDGSIDYLEFISATMHMNRIEREDHLYTAFQFFDNDNSGYITMEELELAMKKYNMGD 487
Query 175 SYGGVLAEEILQQCDIKKNGVIDFEEFMVAL 205
+EI+ + D ++G I++EEF+ +
Sbjct 488 DKS---IKEIIAEVDTDRDGKINYEEFVAMM 515
Score = 33.5 bits (75), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
++E F+ LD D +G ++L+ LR L G ++E ++++ D+ +G ID+ EF+
Sbjct 384 LKEMFKSLDTDNNGIVTLEELRTGL-PKLGSKISEAEIRQLMEAADMDGDGSIDYLEFIS 442
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 443 ATMHMNRIERE 453
> At4g04720
Length=531
Score = 110 bits (274), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 66/213 (30%), Positives = 112/213 (52%), Gaps = 5/213 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L ++P+ R+TAA LEHPW ++ G ID VL M++F A +++
Sbjct 311 AKDLVRKMLTKDPKRRITAAQVLEHPW--IKGGEAPDKPIDSAVLSRMKQFRAMNKLKKL 368
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA S+ +++ L+ +F +D K+G+I E L L + + + E E ++
Sbjct 369 ALKVIAESLSEEEIKGLKTMFANIDTDKSGTITYEELKTGLTRLGSRLSETEVKQLMEAA 428
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGGV 179
D+ G+ I+Y EF++AT + +AF+ D D SG I+ D L + + G
Sbjct 429 DVDGNGTIDYYEFISATMHRYKLDRDEHVYKAFQHFDKDNSGHITRDELESAMKEYGMGD 488
Query 180 LA--EEILQQCDIKKNGVIDFEEFMVALTGAVT 210
A +E++ + D +G I+FEEF + T
Sbjct 489 EASIKEVISEVDTDNDGRINFEEFCAMMRSGST 521
Score = 37.0 bits (84), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query 148 IREAFERLDVDQSGEISLDNLRHIL---GDSYGGVLAEEILQQCDIKKNGVIDFEEFMVA 204
++ F +D D+SG I+ + L+ L G +++++ D+ NG ID+ EF+ A
Sbjct 385 LKTMFANIDTDKSGTITYEELKTGLTRLGSRLSETEVKQLMEAADVDGNGTIDYYEFISA 444
Query 205 LTGAVTFDRE 214
DR+
Sbjct 445 TMHRYKLDRD 454
> At4g04740
Length=520
Score = 107 bits (267), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 65/213 (30%), Positives = 113/213 (53%), Gaps = 5/213 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L +P+ R+TAA LEHPW ++ G ID VL M++F A +++
Sbjct 300 AKDLVEKMLTEDPKRRITAAQVLEHPW--IKGGEAPEKPIDSTVLSRMKQFRAMNKLKKL 357
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL + A+S+ +++ L+ LF +D ++G+I E L L + + + E E ++
Sbjct 358 ALKVSAVSLSEEEIKGLKTLFANMDTNRSGTITYEQLQTGLSRLRSRLSETEVQQLVEAS 417
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGGV 179
D+ G+ I+Y EF++AT + +AF+ LD D++G I+ D L + + G
Sbjct 418 DVDGNGTIDYYEFISATMHRYKLHHDEHVHKAFQHLDKDKNGHITRDELESAMKEYGMGD 477
Query 180 LA--EEILQQCDIKKNGVIDFEEFMVALTGAVT 210
A +E++ + D +G I+FEEF + T
Sbjct 478 EASIKEVISEVDTDNDGKINFEEFRAMMRCGTT 510
> At3g20410
Length=541
Score = 107 bits (267), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 67/209 (32%), Positives = 110/209 (52%), Gaps = 6/209 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L +P+ R++AA L+HPWL E G ID VL M++F A +++
Sbjct 322 AKDLVRRMLTADPKRRISAADVLQHPWLR-EGGEASDKPIDSAVLSRMKQFRAMNKLKKL 380
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFHRI 119
AL +IA ++ ++++ L+ +F +D +G+I E L L K + EAE ++
Sbjct 381 ALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKEGLAKLGSKLTEAEVKQLMDAA 440
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ G+ I+Y EF+ AT S + +AF+ D D SG I++D L L + YG
Sbjct 441 DVDGNGSIDYIEFITATMHRHRLESNENLYKAFQHFDKDSSGYITIDELESALKE-YGMG 499
Query 178 -GVLAEEILQQCDIKKNGVIDFEEFMVAL 205
+E+L D +G I++EEF +
Sbjct 500 DDATIKEVLSDVDSDNDGRINYEEFCAMM 528
> At1g50700
Length=521
Score = 105 bits (263), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 67/209 (32%), Positives = 111/209 (53%), Gaps = 6/209 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L ++P+ R++AA L+HPWL E G ID VL M++F A +++
Sbjct 304 AKDLVRRMLTQDPKRRISAAEVLKHPWLR-EGGEASDKPIDSAVLSRMKQFRAMNKLKKL 362
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA ++ ++++ L+ +F +D +G+I E L L K + + EAE ++
Sbjct 363 ALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKEGLAKLGSRLTEAEVKQLMDAA 422
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ G+ I+Y EF+ AT S + +AF+ D D SG I+ D L L + YG
Sbjct 423 DVDGNGSIDYIEFITATMHRHRLESNENVYKAFQHFDKDGSGYITTDELEAALKE-YGMG 481
Query 178 -GVLAEEILQQCDIKKNGVIDFEEFMVAL 205
+EIL D +G I+++EF +
Sbjct 482 DDATIKEILSDVDADNDGRINYDEFCAMM 510
> At1g76040
Length=534
Score = 105 bits (263), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 67/214 (31%), Positives = 116/214 (54%), Gaps = 7/214 (3%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L R+P+ R+TAA ALEHPW+ D I I+ VL M++F A +++
Sbjct 316 AKDLIRKMLIRDPKKRITAAEALEHPWMT--DTKISDKPINSAVLVRMKQFRAMNKLKKL 373
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA ++ +++ L++ F +D ++G+I + L N L + + + E+E ++
Sbjct 374 ALKVIAENLSEEEIKGLKQTFKNMDTDESGTITFDELRNGLHRLGSKLTESEIKQLMEAA 433
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ I+Y EF+ AT + + EAF+ D D+SG I+ D L+H + + YG
Sbjct 434 DVDKSGTIDYIEFVTATMHRHRLEKEENLIEAFKYFDKDRSGFITRDELKHSMTE-YGMG 492
Query 178 -GVLAEEILQQCDIKKNGVIDFEEFMVALTGAVT 210
+E++ D +G I++EEF+ + T
Sbjct 493 DDATIDEVINDVDTDNDGRINYEEFVAMMRKGTT 526
> At4g35310
Length=556
Score = 102 bits (253), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 66/208 (31%), Positives = 110/208 (52%), Gaps = 11/208 (5%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L P +RLTA L HPW+ E+G +D VL +++F+A +++
Sbjct 328 AKDLIRRMLSSKPAERLTAHEVLRHPWIC-ENGVAPDRALDPAVLSRLKQFSAMNKLKKM 386
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA S+ +++ L ++F A+D +G+I + L L K + + + E H +
Sbjct 387 ALKVIAESLSEEEIAGLREMFQAMDTDNSGAITFDELKAGLRKYGSTLKDTEIHDLMDAA 446
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLR-----HILGD 174
D+ I+YSEF+AAT + + AF+ D D SG I++D L+ H + D
Sbjct 447 DVDNSGTIDYSEFIAATIHLNKLEREEHLVAAFQYFDKDGSGFITIDELQQACVEHGMAD 506
Query 175 SYGGVLAEEILQQCDIKKNGVIDFEEFM 202
V E+I+++ D +G ID+ EF+
Sbjct 507 ----VFLEDIIKEVDQNNDGKIDYGEFV 530
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
+RE F+ +D D SG I+ D L+ L YG L + +++ D+ +G ID+ EF+
Sbjct 403 LREMFQAMDTDNSGAITFDELKAGL-RKYGSTLKDTEIHDLMDAADVDNSGTIDYSEFIA 461
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 462 ATIHLNKLERE 472
> At2g17890
Length=571
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 71/209 (33%), Positives = 106/209 (50%), Gaps = 12/209 (5%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKDF+ LL ++P RLTAA AL HPW+ E G IDI VL +MR+F + +++
Sbjct 341 AKDFVKKLLVKDPRARLTAAQALSHPWVR-EGGDASEIPIDISVLNNMRQFVKFSRLKQF 399
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK--TLNIPEAEAHRIFHR 118
AL +A ++ ++L +L F A+D K G I +E + L K + +A I
Sbjct 400 ALRALATTLDEEELADLRDQFDAIDVDKNGVISLEEMRQALAKDHPWKLKDARVAEILQA 459
Query 119 IDLTGDQEINYSEFLAATFQTQ------VALSQSLIREAFERLDVDQSGEISLDNLRHIL 172
ID D +++ EF+AA Q R AFE+ D+D G I+ + LR
Sbjct 460 IDSNTDGFVDFGEFVAAALHVNQLEEHDSEKWQQRSRAAFEKFDIDGDGFITAEELRMHT 519
Query 173 GDSYGGVLAEEILQQCDIKKNGVIDFEEF 201
G G + E +L++ DI +G I +EF
Sbjct 520 GLK-GSI--EPLLEEADIDNDGKISLQEF 545
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
+R+ F+ +DVD++G ISL+ +R L + L + EILQ D +G +DF EF+
Sbjct 416 LRDQFDAIDVDKNGVISLEEMRQALAKDHPWKLKDARVAEILQAIDSNTDGFVDFGEFVA 475
Query 204 A 204
A
Sbjct 476 A 476
> At5g12180
Length=528
Score = 101 bits (252), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 63/209 (30%), Positives = 110/209 (52%), Gaps = 6/209 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L +P+ RLTAA L HPW+ EDG +D V+ +++F A ++
Sbjct 304 AKDLVKKMLNSDPKQRLTAAQVLNHPWIK-EDGEAPDVPLDNAVMSRLKQFKAMNNFKKV 362
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFHRI 119
AL +IA + +++ L+++F +D +G+I +E L L K + E E ++
Sbjct 363 ALRVIAGCLSEEEIMGLKEMFKGMDTDSSGTITLEELRQGLAKQGTRLSEYEVQQLMEAA 422
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D G+ I+Y EF+AAT + + AF+ D D SG I+++ L L + +G
Sbjct 423 DADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYITMEELEQALRE-FGMN 481
Query 178 -GVLAEEILQQCDIKKNGVIDFEEFMVAL 205
G +EI+ + D +G I+++EF+ +
Sbjct 482 DGRDIKEIISEVDGDNDGRINYDEFVAMM 510
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
++E F+ +D D SG I+L+ LR L G L+E ++++ D NG ID+ EF+
Sbjct 379 LKEMFKGMDTDSSGTITLEELRQGLAKQ-GTRLSEYEVQQLMEAADADGNGTIDYGEFIA 437
Query 204 ALTGAVTFDRE 214
A DRE
Sbjct 438 ATMHINRLDRE 448
> At5g19360
Length=523
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 64/209 (30%), Positives = 109/209 (52%), Gaps = 6/209 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L +P+ RLTAA L HPW+ EDG +D V+ +++F A ++
Sbjct 299 AKDLVRKMLNSDPKQRLTAAQVLNHPWIK-EDGEAPDVPLDNAVMSRLKQFKAMNNFKKV 357
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFHRI 119
AL +IA + +++ L+++F +D +G+I +E L L K + E E ++
Sbjct 358 ALRVIAGCLSEEEIMGLKEMFKGMDTDNSGTITLEELRQGLAKQGTRLSEYEVQQLMEAA 417
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D G+ I+Y EF+AAT + + AF+ D D SG I+ + L L + +G
Sbjct 418 DADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYITTEELEQALRE-FGMN 476
Query 178 -GVLAEEILQQCDIKKNGVIDFEEFMVAL 205
G +EI+ + D +G I++EEF+ +
Sbjct 477 DGRDIKEIISEVDGDNDGRINYEEFVAMM 505
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
++E F+ +D D SG I+L+ LR L G L+E ++++ D NG ID+ EF+
Sbjct 374 LKEMFKGMDTDNSGTITLEELRQGLAKQ-GTRLSEYEVQQLMEAADADGNGTIDYGEFIA 432
Query 204 ALTGAVTFDRE 214
A DRE
Sbjct 433 ATMHINRLDRE 443
> At5g04870
Length=610
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 69/214 (32%), Positives = 112/214 (52%), Gaps = 6/214 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L R+P+ RLTA L HPW+ V DG +D VL M++F+A ++
Sbjct 381 AKDLVRKMLVRDPKKRLTAHQVLCHPWVQV-DGVAPDKPLDSAVLSRMKQFSAMNKFKKM 439
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKT-LNIPEAEAHRIFHRI 119
AL +IA S+ +++ L+++F +D K+G I E L L + N+ E+E +
Sbjct 440 ALRVIAESLSEEEIAGLKEMFNMIDADKSGQITFEELKAGLKRVGANLKESEILDLMQAA 499
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ I+Y EF+AAT + + AF D D SG I+ D L+ + +G
Sbjct 500 DVDNSGTIDYKEFIAATLHLNKIEREDHLFAAFTYFDKDGSGYITPDELQQAC-EEFGVE 558
Query 178 GVLAEEILQQCDIKKNGVIDFEEFMVAL-TGAVT 210
V EE+++ D +G ID+ EF+ + G++T
Sbjct 559 DVRIEELMRDVDQDNDGRIDYNEFVAMMQKGSIT 592
> At1g61950
Length=547
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 61/203 (30%), Positives = 109/203 (53%), Gaps = 6/203 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + ++L+ +P+ R TAA LEHPW+ E G ID VL M++ A +++
Sbjct 330 AKDLVRNMLKYDPKKRFTAAQVLEHPWIR-EGGEASDKPIDSAVLSRMKQLRAMNKLKKL 388
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
A IA ++ ++L L+ +F +D K+G+I + L + L K + + E E ++
Sbjct 389 AFKFIAQNLKEEELKGLKTMFANMDTDKSGTITYDELKSGLEKLGSRLTETEVKQLLEDA 448
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSG-EISLDNLRHILGDSYGG 178
D+ G+ I+Y EF++AT + + +AF+ D D SG E+ + +GD
Sbjct 449 DVDGNGTIDYIEFISATMNRFRVEREDNLFKAFQHFDKDNSGQELETAMKEYNMGDD--- 505
Query 179 VLAEEILQQCDIKKNGVIDFEEF 201
++ +EI+ + D +G I+++EF
Sbjct 506 IMIKEIISEVDADNDGSINYQEF 528
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
++ F +D D+SG I+ D L+ L + G L E ++L+ D+ NG ID+ EF+
Sbjct 405 LKTMFANMDTDKSGTITYDELKSGL-EKLGSRLTETEVKQLLEDADVDGNGTIDYIEFIS 463
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 464 ATMNRFRVERE 474
> At2g17290
Length=544
Score = 99.8 bits (247), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 65/211 (30%), Positives = 111/211 (52%), Gaps = 11/211 (5%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L +P +RLTA L HPW+ E+G +D VL +++F+A +++
Sbjct 316 AKDLIRKMLCSSPSERLTAHEVLRHPWIC-ENGVAPDRALDPAVLSRLKQFSAMNKLKKM 374
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA S+ +++ L +F A+D +G+I + L L + + + + E +
Sbjct 375 ALKVIAESLSEEEIAGLRAMFEAMDTDNSGAITFDELKAGLRRYGSTLKDTEIRDLMEAA 434
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLR-----HILGD 174
D+ I+YSEF+AAT + + AF+ D D SG I++D L+ H + D
Sbjct 435 DVDNSGTIDYSEFIAATIHLNKLEREEHLVSAFQYFDKDGSGYITIDELQQSCIEHGMTD 494
Query 175 SYGGVLAEEILQQCDIKKNGVIDFEEFMVAL 205
V E+I+++ D +G ID+EEF+ +
Sbjct 495 ----VFLEDIIKEVDQDNDGRIDYEEFVAMM 521
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
+R FE +D D SG I+ D L+ L YG L + ++++ D+ +G ID+ EF+
Sbjct 391 LRAMFEAMDTDNSGAITFDELKAGL-RRYGSTLKDTEIRDLMEAADVDNSGTIDYSEFIA 449
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 450 ATIHLNKLERE 460
> At3g10660
Length=646
Score = 99.0 bits (245), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 65/208 (31%), Positives = 108/208 (51%), Gaps = 5/208 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L R+P+ RLTA L HPW+ + DG +D VL M++F+A ++
Sbjct 417 AKDLVRKMLVRDPKRRLTAHQVLCHPWVQI-DGVAPDKPLDSAVLSRMKQFSAMNKFKKM 475
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKT-LNIPEAEAHRIFHRI 119
AL +IA S+ +++ L+++F +D +G I E L L + N+ E+E +
Sbjct 476 ALRVIAESLSEEEIAGLKQMFKMIDADNSGQITFEELKAGLKRVGANLKESEILDLMQAA 535
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ I+Y EF+AAT + + AF D D+SG I+ D L+ + +G
Sbjct 536 DVDNSGTIDYKEFIAATLHLNKIEREDHLFAAFSYFDKDESGFITPDELQQAC-EEFGVE 594
Query 178 GVLAEEILQQCDIKKNGVIDFEEFMVAL 205
EE+++ D K+G ID+ EF+ +
Sbjct 595 DARIEEMMRDVDQDKDGRIDYNEFVAMM 622
Score = 37.7 bits (86), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
+++ F+ +D D SG+I+ + L+ L G L E +++Q D+ +G ID++EF+
Sbjct 492 LKQMFKMIDADNSGQITFEELKAGL-KRVGANLKESEILDLMQAADVDNSGTIDYKEFIA 550
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 551 ATLHLNKIERE 561
> At4g38230
Length=484
Score = 95.9 bits (237), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 60/207 (28%), Positives = 108/207 (52%), Gaps = 3/207 (1%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AK+ I +L P +RLTA L HPW+ E+G +D VL +++F+A +++
Sbjct 255 AKNLIRGMLCSRPSERLTAHQVLRHPWIC-ENGVAPDRALDPAVLSRLKQFSAMNKLKQM 313
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA S+ +++ L+++F A+D +G+I + L L + + + + E +
Sbjct 314 ALRVIAESLSEEEIAGLKEMFKAMDTDNSGAITFDELKAGLRRYGSTLKDTEIRDLMEAA 373
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGD-SYGG 178
D+ I+Y EF+AAT + + AF D D SG I++D L+H +
Sbjct 374 DIDKSGTIDYGEFIAATIHLNKLEREEHLLSAFRYFDKDGSGYITIDELQHACAEQGMSD 433
Query 179 VLAEEILQQCDIKKNGVIDFEEFMVAL 205
V E+++++ D +G ID+ EF+ +
Sbjct 434 VFLEDVIKEVDQDNDGRIDYGEFVAMM 460
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
++E F+ +D D SG I+ D L+ L YG L + ++++ DI K+G ID+ EF+
Sbjct 330 LKEMFKAMDTDNSGAITFDELKAGL-RRYGSTLKDTEIRDLMEAADIDKSGTIDYGEFIA 388
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 389 ATIHLNKLERE 399
> At4g04710
Length=575
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 72/250 (28%), Positives = 122/250 (48%), Gaps = 29/250 (11%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AK I +L + P++R++AA LEHPW+ E ID VL M++F A +++
Sbjct 264 AKHLIGKMLTKKPKERISAADVLEHPWMKSE---APDKPIDNVVLSRMKQFRAMNKLKKL 320
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFHRI 119
AL +IA + +++ L+ +F +D K+GSI E L L + + E E ++ +
Sbjct 321 ALKVIAEGLSEEEIKGLKTMFENMDMDKSGSITYEELKMGLNRHGSKLSETEVKQLMEAV 380
Query 120 --DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNL-----RHIL 172
D+ G+ I+Y EF++AT + +AF+ D D SG I+ + + H +
Sbjct 381 SADVDGNGTIDYIEFISATMHRHRLERDEHLYKAFQYFDKDGSGHITKEEVEIAMKEHGM 440
Query 173 GDSYGGVLAEEILQQCDIKKNGVIDFEEFMVALTGAVTFDRERGNSFLQKLPRKEQSEVL 232
GD A++++ + D +G ID+EEF + N LQ Q ++L
Sbjct 441 GDEAN---AKDLISEFDKNNDGKIDYEEFCTMMR----------NGILQP-----QGKLL 482
Query 233 SRVQRNLSDL 242
R+ NL +L
Sbjct 483 KRLYMNLEEL 492
> At5g66210
Length=523
Score = 93.6 bits (231), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 66/209 (31%), Positives = 107/209 (51%), Gaps = 12/209 (5%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKDF+ LL ++P RLTAA AL H W+ E G+ +DI VL ++R+F + +++
Sbjct 295 AKDFVKKLLVKDPRARLTAAQALSHAWVR-EGGNATDIPVDISVLNNLRQFVRYSRLKQF 353
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHR 118
AL +A ++ ++ +L F A+D K G I +E + L K L + ++ I
Sbjct 354 ALRALASTLDEAEISDLRDQFDAIDVDKNGVISLEEMRQALAKDLPWKLKDSRVAEILEA 413
Query 119 IDLTGDQEINYSEFLAATFQTQ------VALSQSLIREAFERLDVDQSGEISLDNLRHIL 172
ID D ++++EF+AA Q R AFE+ D+D+ G I+ + LR
Sbjct 414 IDSNTDGLVDFTEFVAAALHVHQLEEHDSEKWQLRSRAAFEKFDLDKDGYITPEELRMHT 473
Query 173 GDSYGGVLAEEILQQCDIKKNGVIDFEEF 201
G G + + +L + DI ++G I EF
Sbjct 474 G-LRGSI--DPLLDEADIDRDGKISLHEF 499
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
+R+ F+ +DVD++G ISL+ +R L L + EIL+ D +G++DF EF+
Sbjct 370 LRDQFDAIDVDKNGVISLEEMRQALAKDLPWKLKDSRVAEILEAIDSNTDGLVDFTEFVA 429
Query 204 A 204
A
Sbjct 430 A 430
> At1g35670
Length=495
Score = 92.4 bits (228), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 63/218 (28%), Positives = 111/218 (50%), Gaps = 5/218 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L R+P+ R++A AL HPW+ E + + +D VL +++F+ I++
Sbjct 257 AKDLIYKMLERSPKKRISAHEALCHPWIVDEQAAPDK-PLDPAVLSRLKQFSQMNKIKKM 315
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA + +++ L++LF +D +G+I E L L + + + E+E +
Sbjct 316 ALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKRVGSELMESEIKSLMDAA 375
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ I+Y EFLAAT + + AF D D SG I++D L+ + +G
Sbjct 376 DIDNSGTIDYGEFLAATLHMNKMEREENLVAAFSYFDKDGSGYITIDELQSACTE-FGLC 434
Query 178 GVLAEEILQQCDIKKNGVIDFEEFMVALTGAVTFDRER 215
++++++ D+ +G IDF EF + R R
Sbjct 435 DTPLDDMIKEIDLDNDGKIDFSEFTAMMRKGDGVGRSR 472
> At4g09570
Length=501
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 63/218 (28%), Positives = 112/218 (51%), Gaps = 5/218 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L R+P+ R++A AL HPW+ V++ + +D VL +++F+ I++
Sbjct 256 AKDLIYKMLDRSPKKRISAHEALCHPWI-VDEHAAPDKPLDPAVLSRLKQFSQMNKIKKM 314
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA + +++ L++LF +D +G+I E L L + + + E+E +
Sbjct 315 ALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKRVGSELMESEIKSLMDAA 374
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ I+Y EFLAAT + + AF D D SG I++D L+ + +G
Sbjct 375 DIDNSGTIDYGEFLAATLHINKMEREENLVVAFSYFDKDGSGYITIDELQQACTE-FGLC 433
Query 178 GVLAEEILQQCDIKKNGVIDFEEFMVALTGAVTFDRER 215
++++++ D+ +G IDF EF + R R
Sbjct 434 DTPLDDMIKEIDLDNDGKIDFSEFTAMMKKGDGVGRSR 471
> At4g36070
Length=536
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 67/211 (31%), Positives = 104/211 (49%), Gaps = 11/211 (5%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKDF+ LL + P RLTAA AL H W+ E G IDI VL +MR+F + +++
Sbjct 301 AKDFVKKLLVKEPRARLTAAQALSHSWVK-EGGEASEVPIDISVLNNMRQFVKFSRLKQI 359
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHR 118
AL +A ++ +LD+L F A+D K GSI +E + L K + + +A I
Sbjct 360 ALRALAKTINEDELDDLRDQFDAIDIDKNGSISLEEMRQALAKDVPWKLKDARVAEILQA 419
Query 119 IDLTGDQEINYSEFLAATFQTQ------VALSQSLIREAFERLDVDQSGEISLDNLR--H 170
D D ++++EF+ A Q R AF++ D+D G I+ + LR
Sbjct 420 NDSNTDGLVDFTEFVVAALHVNQLEEHDSEKWQQRSRAAFDKFDIDGDGFITPEELRLNQ 479
Query 171 ILGDSYGGVLAEEILQQCDIKKNGVIDFEEF 201
L + E +L++ D+ ++G I EF
Sbjct 480 CLQQTGLKGSIEPLLEEADVDEDGRISINEF 510
> At5g19450
Length=533
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 68/238 (28%), Positives = 117/238 (49%), Gaps = 15/238 (6%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKS-MRRFAACTAIQR 59
AKD + +L +P+ RL+AA LEH W +++ + E +K+ +++F+ +++
Sbjct 288 AKDLVRKMLEPDPKKRLSAAQVLEHSW--IQNAKKAPNVSLGETVKARLKQFSVMNKLKK 345
Query 60 AALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN--IPEAEAHRIFH 117
AL +IA + +++ +++ F +D KKTG I +E L L K IP+ + +
Sbjct 346 RALRVIAEHLSVEEVAGIKEAFEMMDSKKTGKINLEELKFGLHKLGQQQIPDTDLQILME 405
Query 118 RIDLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILG---D 174
D+ GD +NY EF+A + + + + +AF D +QS I ++ LR L D
Sbjct 406 AADVDGDGTLNYGEFVAVSVHLKKMANDEHLHKAFSFFDQNQSDYIEIEELREALNDEVD 465
Query 175 SYGGVLAEEILQQCDIKKNGVIDFEEFMVALTGAVT-------FDRERGNSFLQKLPR 225
+ + I+Q D K+G I +EEF + + RER NS KL R
Sbjct 466 TNSEEVVAAIMQDVDTDKDGRISYEEFAAMMKAGTDWRKASRQYSRERFNSLSLKLMR 523
> At5g23580
Length=490
Score = 90.5 bits (223), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 62/211 (29%), Positives = 109/211 (51%), Gaps = 11/211 (5%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L NP+ RLTA L HPW+ V+D +D V+ +++F+A +++
Sbjct 253 AKDLIKKMLESNPKKRLTAHQVLCHPWI-VDDKVAPDKPLDCAVVSRLKKFSAMNKLKKM 311
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA + +++ L++LF +D K+G+I E L + + + + + E+E +
Sbjct 312 ALRVIAERLSEEEIGGLKELFKMIDTDKSGTITFEELKDSMRRVGSELMESEIQELLRAA 371
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHI-----LGD 174
D+ I+Y EFLAAT + + AF D D SG I+++ L+ + D
Sbjct 372 DVDESGTIDYGEFLAATIHLNKLEREENLVAAFSFFDKDASGYITIEELQQAWKEFGIND 431
Query 175 SYGGVLAEEILQQCDIKKNGVIDFEEFMVAL 205
S +E+++ D +G ID+ EF+ +
Sbjct 432 SN----LDEMIKDIDQDNDGQIDYGEFVAMM 458
Score = 36.6 bits (83), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAE----EILQQCDIKKNGVIDFEEFMV 203
++E F+ +D D+SG I+ + L+ + G L E E+L+ D+ ++G ID+ EF+
Sbjct 328 LKELFKMIDTDKSGTITFEELKDSM-RRVGSELMESEIQELLRAADVDESGTIDYGEFLA 386
Query 204 ALTGAVTFDRE 214
A +RE
Sbjct 387 ATIHLNKLERE 397
> At2g38910
Length=583
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 59/205 (28%), Positives = 105/205 (51%), Gaps = 5/205 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD + +L R+P+ R+T L HPW V DG +D VL +++F+A +++
Sbjct 365 AKDLVRRMLIRDPKKRMTTHEVLCHPWARV-DGVALDKPLDSAVLSRLQQFSAMNKLKKI 423
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKT-LNIPEAEAHRIFHRI 119
A+ +IA S+ +++ L+++F +D +G I +E L L + ++ ++E +
Sbjct 424 AIKVIAESLSEEEIAGLKEMFKMIDTDNSGHITLEELKKGLDRVGADLKDSEILGLMQAA 483
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ I+Y EF+AA + + AF D D SG I+ D L+ +G
Sbjct 484 DIDNSGTIDYGEFIAAMVHLNKIEKEDHLFTAFSYFDQDGSGYITRDELQQAC-KQFGLA 542
Query 178 GVLAEEILQQCDIKKNGVIDFEEFM 202
V ++IL++ D +G ID+ EF+
Sbjct 543 DVHLDDILREVDKDNDGRIDYSEFV 567
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 36/63 (57%), Gaps = 5/63 (7%)
Query 148 IREAFERLDVDQSGEISLDNLRHILGDSYGGVLAEE----ILQQCDIKKNGVIDFEEFMV 203
++E F+ +D D SG I+L+ L+ L D G L + ++Q DI +G ID+ EF+
Sbjct 440 LKEMFKMIDTDNSGHITLEELKKGL-DRVGADLKDSEILGLMQAADIDNSGTIDYGEFIA 498
Query 204 ALT 206
A+
Sbjct 499 AMV 501
> At1g18890
Length=545
Score = 80.9 bits (198), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 57/215 (26%), Positives = 101/215 (46%), Gaps = 5/215 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AK + +L +P RLTA L HPW+ + DI V +++F+ ++
Sbjct 294 AKSLVKQMLDPDPTKRLTAQQVLAHPWIQNAKKAPNVPLGDI-VRSRLKQFSMMNRFKKK 352
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
L +IA + ++++ ++ +F +D+ K G I L L K + + E E +
Sbjct 353 VLRVIAEHLSIQEVEVIKNMFSLMDDDKDGKITYPELKAGLQKVGSQLGEPEIKMLMEVA 412
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG-- 177
D+ G+ ++Y EF+A Q + L + AF D D S I LD LR L D G
Sbjct 413 DVDGNGFLDYGEFVAVIIHLQKIENDELFKLAFMFFDKDGSTYIELDELREALADELGEP 472
Query 178 -GVLAEEILQQCDIKKNGVIDFEEFMVALTGAVTF 211
+ +I+++ D K+G I+++EF+ + +
Sbjct 473 DASVLSDIMREVDTDKDGRINYDEFVTMMKAGTDW 507
> At2g31500
Length=582
Score = 73.2 bits (178), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 52/218 (23%), Positives = 102/218 (46%), Gaps = 8/218 (3%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AK+ + ++L NP RLT LEHPW+ + + + D V +++F ++
Sbjct 297 AKELVKNMLDANPYSRLTVQEVLEHPWIRNAERAPNVNLGD-NVRTKIQQFLLMNRFKKK 355
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
L ++A ++P++++ + ++F +D K G + E L + L K +P+ + +
Sbjct 356 VLRIVADNLPNEEIAAIVQMFQTMDTDKNGHLTFEELRDGLKKIGQVVPDGDVKMLMDAA 415
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGGV 179
D G+ ++ EF+ + + ++EAF+ D + +G I LD L+ L D G
Sbjct 416 DTDGNGMLSCDEFVTLSIHLKRMGCDEHLQEAFKYFDKNGNGFIELDELKVALCDDKLGH 475
Query 180 ------LAEEILQQCDIKKNGVIDFEEFMVALTGAVTF 211
++I D+ K+G I F+EF + +
Sbjct 476 ANGNDQWIKDIFFDVDLNKDGRISFDEFKAMMKSGTDW 513
> At3g51850
Length=503
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 59/232 (25%), Positives = 110/232 (47%), Gaps = 33/232 (14%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKS-MRRFAACTAIQR 59
AK+ + +L +P+ RLTA LEHPW +++ + +V+KS +++F+ +R
Sbjct 285 AKNLVRQMLEPDPKRRLTAKQVLEHPW--IQNAKKAPNVPLGDVVKSRLKQFSVMNRFKR 342
Query 60 AALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFHR 118
AL +IA + ++++++++ +F +D G + +E L L + + E+E +
Sbjct 343 KALRVIAEFLSTEEVEDIKVMFNKMDTDNDGIVSIEELKAGLRDFSTQLAESEVQMLIEA 402
Query 119 IDLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGG 178
+D G ++Y EF+A + Q L +L + G+ +D
Sbjct 403 VDTKGKGTLDYGEFVAVSLHLQKELCDALKEDG---------GDDCVD------------ 441
Query 179 VLAEEILQQCDIKKNGVIDFEEFMVAL-TG------AVTFDRERGNSFLQKL 223
+A +I Q+ D K+G I +EEF + TG + + R R NS KL
Sbjct 442 -VANDIFQEVDTDKDGRISYEEFAAMMKTGTDWRKASRHYSRGRFNSLSIKL 492
> At2g41860
Length=530
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 54/214 (25%), Positives = 106/214 (49%), Gaps = 8/214 (3%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKS-MRRFAACTAIQR 59
AKD I +L +P RLTA L+HPW +++G + E +++ +++F+ +++
Sbjct 285 AKDLIKKMLHPDPRRRLTAQQVLDHPW--IQNGKNASNVSLGETVRARLKQFSVMNKLKK 342
Query 60 AALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFHR 118
AL +IA + ++ +++ F +D G I + L L K + +P+ + +
Sbjct 343 RALRVIAEHLSVEETSCIKERFQVMDTSNRGKITITELGIGLQKLGIVVPQDDIQILMDA 402
Query 119 IDLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGG 178
D+ D ++ +EF+A + + + +++AF D ++SG I ++ LR L D
Sbjct 403 GDVDKDGYLDVNEFVAISVHIRKLGNDEHLKKAFTFFDKNKSGYIEIEELRDALADDVDT 462
Query 179 V---LAEEILQQCDIKKNGVIDFEEFMVAL-TGA 208
+ E I+ D K+G I ++EF + TG
Sbjct 463 TSEEVVEAIILDVDTNKDGKISYDEFATMMKTGT 496
> At3g57530
Length=560
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 59/233 (25%), Positives = 109/233 (46%), Gaps = 11/233 (4%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L + + RLTA L+HPWL + ++ V +++F +++
Sbjct 294 AKDLIRKMLDPDQKRRLTAQQVLDHPWLQ-NAKTAPNVSLGETVRARLKQFTVMNKLKKR 352
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRI 119
AL +IA + ++ + + F +D + G I ++ L L K + IP+ + +
Sbjct 353 ALRVIAEHLSDEEASGIREGFQIMDTSQRGKINIDELKIGLQKLGHAIPQDDLQILMDAG 412
Query 120 DLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGGV 179
D+ D ++ EF+A + + + +++AF D + +G I ++ LR L D G
Sbjct 413 DIDRDGYLDCDEFIAISVHLRKMGNDEHLKKAFAFFDQNNNGYIEIEELREALSDELGTS 472
Query 180 --LAEEILQQCDIKKNGVIDFEEFMVALTGAVT-------FDRERGNSFLQKL 223
+ + I++ D K+G I +EEF+ + + RER NS KL
Sbjct 473 EEVVDAIIRDVDTDKDGRISYEEFVTMMKTGTDWRKASRQYSRERFNSISLKL 525
> At2g35890
Length=520
Score = 70.1 bits (170), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 48/158 (30%), Positives = 85/158 (53%), Gaps = 7/158 (4%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKD I +L RNP RLTA L HPW+ ++G+ +D VL +++F+A +++
Sbjct 363 AKDLIRKMLERNPIQRLTAQQVLCHPWIR-DEGNAPDTPLDTTVLSRLKKFSATDKLKKM 421
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK-TLNIPEAEAHRIFH-R 118
AL +IA + +++ EL + F +D K+G + + L N L + N+ ++ + +
Sbjct 422 ALRVIAERLSEEEIHELRETFKTIDSGKSGRVTYKELKNGLERFNTNLDNSDINSLMQIP 481
Query 119 IDLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLD 156
D+ + ++Y+EF+ A V L Q EA +RL+
Sbjct 482 TDVHLEDTVDYNEFIEAI----VRLRQIQEEEANDRLE 515
> At3g50530
Length=601
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 56/213 (26%), Positives = 104/213 (48%), Gaps = 9/213 (4%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
A+DF+ LL ++P RLTAA AL HPW ++D + + +DI V K MR + +++++A
Sbjct 383 ARDFVKRLLNKDPRKRLTAAQALSHPW--IKDSNDAKVPMDILVFKLMRAYLRSSSLRKA 440
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK--TLNIPEAEAHRIFHR 118
AL ++ ++ +L L + F L+ K G+I +E++ + L+K T + ++ +
Sbjct 441 ALRALSKTLTVDELFYLREQFALLEPSKNGTISLENIKSALMKMATDAMKDSRIPEFLGQ 500
Query 119 IDLTGDQEINYSEFLAATFQTQ----VALSQSLIREAFERLDVDQSGEISLDNLRHILGD 174
+ + +++ EF AA + + R A+E + + + I +D L LG
Sbjct 501 LSALQYRRMDFEEFCAAALSVHQLEALDRWEQHARCAYELFEKEGNRPIMIDELASELGL 560
Query 175 SYGGVLAEEILQQCDIKKNGVIDFEEFMVALTG 207
V +L +G + F F+ L G
Sbjct 561 G-PSVPVHAVLHDWLRHTDGKLSFLGFVKLLHG 592
> At1g74740
Length=567
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 48/192 (25%), Positives = 91/192 (47%), Gaps = 5/192 (2%)
Query 24 EHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRAALGLIAMSMPSKDLDELEKLFCA 83
+HPW+ + DI V +++F+ +++ AL +IA + ++++ + +F
Sbjct 339 DHPWIQNAKKAPNVPLGDI-VRSRLKQFSMMNRLKKKALRVIAEHLSIQEVEVIRNMFTL 397
Query 84 LDEKKTGSIRVESLVNVLVKTLN-IPEAEAHRIFHRIDLTGDQEINYSEFLAATFQTQVA 142
+D+ G I L L K + + E E + D+ G+ ++Y EF+A Q
Sbjct 398 MDDDNDGKISYLELRAGLRKVGSQLGEPEIKLLMEVADVNGNGCLDYGEFVAVIIHLQKM 457
Query 143 LSQSLIREAFERLDVDQSGEISLDNLRHILGDSYG---GVLAEEILQQCDIKKNGVIDFE 199
+ R+AF D D SG I + LR L D G + +I+++ D K+G I+++
Sbjct 458 ENDEHFRQAFMFFDKDGSGYIESEELREALTDELGEPDNSVIIDIMREVDTDKDGKINYD 517
Query 200 EFMVALTGAVTF 211
EF+V + +
Sbjct 518 EFVVMMKAGTDW 529
> CE02445
Length=229
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 76/144 (52%), Gaps = 8/144 (5%)
Query 65 IAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKT-LNIPEAEAHRIFHRIDLTG 123
I M K+L E +LF D +G+I E L ++ L+ +AE + +D G
Sbjct 41 IQMKYTRKELKEYRQLFNMFDTDGSGAIGNEELKQAMISIGLHANKAEIDNVIKEVDADG 100
Query 124 DQEINYSEFLAATFQTQ---VALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYG 177
+ EI++ EF A ++Q + ++ LIRE FE D D++G I+ + ++I GD +
Sbjct 101 NGEIDFEEFCACMKKSQNIVKSTNEELIRECFEIFDQDRNGIITENEFKYIAKEFGD-FD 159
Query 178 GVLAEEILQQCDIKKNGVIDFEEF 201
LAE++ ++ D+ NG + ++F
Sbjct 160 DELAEKVFRELDVSANGHLSADQF 183
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 149 REAFERLDVDQSGEISLDNLRHIL---GDSYGGVLAEEILQQCDIKKNGVIDFEEFMVAL 205
R+ F D D SG I + L+ + G + ++++ D NG IDFEEF +
Sbjct 54 RQLFNMFDTDGSGAIGNEELKQAMISIGLHANKAEIDNVIKEVDADGNGEIDFEEFCACM 113
> Hs8393159
Length=146
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 67/128 (52%), Gaps = 8/128 (6%)
Query 79 KLFCALDEKKTGSIRVESLVNVLVKT-LNIPEAEAHRIFHRIDLTGDQEINYSEFLAATF 137
K F A+D G+I + L L T N+ EA+ ++ +D GD EI++ EFL A
Sbjct 15 KAFSAVDTDGNGTINAQELGAALKATGKNLSEAQLRKLISEVDSDGDGEISFQEFLTAAR 74
Query 138 QTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGGVLAEE----ILQQCDIKKN 193
+ + L ++ AF D D G I++D LR + G L +E ++++ D+ ++
Sbjct 75 KARAGLED--LQVAFRAFDQDGDGHITVDELRRAMA-GLGQPLPQEELDAMIREADVDQD 131
Query 194 GVIDFEEF 201
G +++EEF
Sbjct 132 GRVNYEEF 139
> At3g19100
Length=599
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 57/223 (25%), Positives = 106/223 (47%), Gaps = 23/223 (10%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTI--DIEVLKSMRRFAACTAIQ 58
AKDF+ LL ++P R+TA+ AL HPW+A ++ I DI + K ++ + ++++
Sbjct 379 AKDFVKRLLYKDPRKRMTASQALMHPWIA----GYKKIDIPFDILIFKQIKAYLRSSSLR 434
Query 59 RAALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVL-------VKTLNIPEAE 111
+AAL ++ ++ + +L L+ F L K G I ++S+ L +K IP+
Sbjct 435 KAALMALSKTLTTDELLYLKAQFAHLAPNKNGLITLDSIRLALATNATEAMKESRIPDFL 494
Query 112 AHRIFHRIDLTGDQEINYSEFLAATFQTQ----VALSQSLIREAFERLDVDQSGEISLDN 167
A + + + G +++ EF AA+ + + IR A+E +++ + I ++
Sbjct 495 A--LLNGLQYKG---MDFEEFCAASISVHQHESLDCWEQSIRHAYELFEMNGNRVIVIEE 549
Query 168 LRHILGDSYGGVLAEEILQQCDIKKNGVIDFEEFMVALTGAVT 210
L LG + IL +G + F F+ L G T
Sbjct 550 LASELGVG-SSIPVHTILNDWIRHTDGKLSFLGFVKLLHGVST 591
> 7303485
Length=149
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 73/137 (53%), Gaps = 7/137 (5%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFL 133
E ++ F D+ G+I + L V+ ++L N EAE + + +D G+ I++ EFL
Sbjct 12 EFKEAFSLFDKDGDGTITTKELGTVM-RSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 134 AATF-QTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQCD 189
+ + S+ IREAF D D +G IS LRH+ LG+ +E++++ D
Sbjct 71 TMMARKMKDTDSEEEIREAFRVFDKDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREAD 130
Query 190 IKKNGVIDFEEFMVALT 206
I +G +++EEF+ +T
Sbjct 131 IDGDGQVNYEEFVTMMT 147
> CE13902
Length=149
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 73/137 (53%), Gaps = 7/137 (5%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFL 133
E ++ F D+ G+I + L V+ ++L N EAE + + +D G+ I++ EFL
Sbjct 12 EFKEAFSLFDKDGDGTITTKELGTVM-RSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 134 AATF-QTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQCD 189
+ + S+ IREAF D D +G IS LRH+ LG+ +E++++ D
Sbjct 71 TMMARKMKDTDSEEEIREAFRVFDKDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREAD 130
Query 190 IKKNGVIDFEEFMVALT 206
I +G +++EEF+ +T
Sbjct 131 IDGDGQVNYEEFVTMMT 147
> Hs4502549
Length=149
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 73/137 (53%), Gaps = 7/137 (5%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFL 133
E ++ F D+ G+I + L V+ ++L N EAE + + +D G+ I++ EFL
Sbjct 12 EFKEAFSLFDKDGDGTITTKELGTVM-RSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 134 AATF-QTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQCD 189
+ + S+ IREAF D D +G IS LRH+ LG+ +E++++ D
Sbjct 71 TMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREAD 130
Query 190 IKKNGVIDFEEFMVALT 206
I +G +++EEF+ +T
Sbjct 131 IDGDGQVNYEEFVQMMT 147
> At5g24430
Length=594
Score = 59.3 bits (142), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 71/137 (51%), Gaps = 4/137 (2%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKDF+ LL ++ R+TAA AL HPWL E+ + +D V K ++ + + +R+
Sbjct 378 AKDFVKRLLNKDHRKRMTAAQALAHPWLRDENPGL---LLDFSVYKLVKSYIRASPFRRS 434
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTG-SIRVESLVNVLVKTLNIPEAEAHRIFHRI 119
AL ++ ++P ++L L+ F LD K G S+ ++ T + E+ I + +
Sbjct 435 ALKALSKAIPDEELVFLKAQFMLLDPKDGGLSLNCFTMALTRYATDAMMESRLPDILNTM 494
Query 120 DLTGDQEINYSEFLAAT 136
+++++ EF AA
Sbjct 495 QPLAQKKLDFEEFCAAA 511
> At4g14640
Length=151
Score = 59.3 bits (142), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 72/133 (54%), Gaps = 7/133 (5%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFL 133
E ++ FC D+ G I VE L V +++L N E E H I ID + I ++EFL
Sbjct 13 EFKEAFCLFDKDGDGCITVEELATV-IRSLDQNPTEQELHDIITEIDSDSNGTIEFAEFL 71
Query 134 AATFQT-QVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQCD 189
+ Q + ++ ++EAF+ D DQ+G IS L H+ LG+ E+++++ D
Sbjct 72 NLMAKKLQESDAEEELKEAFKVFDKDQNGYISASELSHVMINLGEKLTDEEVEQMIKEAD 131
Query 190 IKKNGVIDFEEFM 202
+ +G ++++EF+
Sbjct 132 LDGDGQVNYDEFV 144
> At3g49370
Length=594
Score = 59.3 bits (142), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 50/213 (23%), Positives = 103/213 (48%), Gaps = 18/213 (8%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKDF+ LL ++ R+TAA AL HPWL E+ + +D + K ++ + + +RA
Sbjct 377 AKDFVKRLLNKDHRKRMTAAQALAHPWLRDENPGL---LLDFSIYKLVKSYIRASPFRRA 433
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVK--TLNIPEAEAHRIFHR 118
AL ++ ++P ++L L+ F L E + G + + + L + T + E+ I +
Sbjct 434 ALKSLSKAIPEEELVFLKAQFMLL-EPEDGGLHLHNFTTALTRYATDAMIESRLPDILNM 492
Query 119 IDLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHILGDSYGG 178
+ +++++ EF AA+ V++ Q EA E + +I+ H +
Sbjct 493 MQPLAHKKLDFEEFCAAS----VSVYQ---LEALEEWE-----QIATVAFEHFESEGSRA 540
Query 179 VLAEEILQQCDIKKNGVIDFEEFMVALTGAVTF 211
+ +E+ ++ + N ++++ +L G + F
Sbjct 541 ISVQELAEEMSLGPNAYPLLKDWIRSLDGKLNF 573
> CE27751
Length=202
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 74/148 (50%), Gaps = 8/148 (5%)
Query 72 KDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLNIPEA--EAHRIFHRIDLTGDQEINY 129
++L E ++F D ++G+I ++ L +K L + + E +I +D G+ +I++
Sbjct 53 EELQEYRQVFNMFDADRSGAIAIDEL-EAAIKNLGLEQTRDELDKIIDEVDQRGNHQIDF 111
Query 130 SEFLAATFQTQVALSQ--SLIREAFERLDVDQSGEISLDNLRHIL---GDSYGGVLAEEI 184
EF + + S +++E F D ++G IS + R IL GD + +EI
Sbjct 112 DEFCVVMRRLTMKKSNWNEVVKECFTVFDRSENGGISKKDFRFILRELGDITDNQIIDEI 171
Query 185 LQQCDIKKNGVIDFEEFMVALTGAVTFD 212
+ D+ NGVID++EF + +T D
Sbjct 172 FNEADVDGNGVIDYDEFTYMVKNYMTDD 199
> At3g22930
Length=173
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 73/140 (52%), Gaps = 9/140 (6%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFL 133
E ++ FC D+ G I + L V +++L N E E + ID G+ I +SEFL
Sbjct 35 EFKEAFCLFDKDGDGCITADELATV-IRSLDQNPTEQELQDMITEIDSDGNGTIEFSEFL 93
Query 134 A-ATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQCD 189
Q Q + ++EAF+ D DQ+G IS LRH+ LG+ ++++++ D
Sbjct 94 NLMANQLQETDADEELKEAFKVFDKDQNGYISASELRHVMINLGEKLTDEEVDQMIKEAD 153
Query 190 IKKNGVIDFEEF--MVALTG 207
+ +G ++++EF M+ + G
Sbjct 154 LDGDGQVNYDEFVRMMMING 173
> At2g41140
Length=576
Score = 57.8 bits (138), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 55/215 (25%), Positives = 100/215 (46%), Gaps = 13/215 (6%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQ-RHTIDIEVLKSMRRFAACTAIQR 59
A DF+ LL ++ RLTAA AL HPWL GS + + D+ + K ++ + T++++
Sbjct 358 AVDFVKRLLNKDYRKRLTAAQALCHPWLV---GSHELKIPSDMIIYKLVKVYIMSTSLRK 414
Query 60 AALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVLVKTLNIPEAEAHRIF--- 116
+AL +A ++ L L + F L K G I +++ ++K+ + + R+F
Sbjct 415 SALAALAKTLTVPQLAYLREQFTLLGPSKNGYISMQNYKTAILKS-STDAMKDSRVFDFV 473
Query 117 HRIDLTGDQEINYSEFLAATFQTQ----VALSQSLIREAFERLDVDQSGEISLDNLRHIL 172
H I +++++ EF A+ + + R A+E + D + I ++ L L
Sbjct 474 HMISCLQYKKLDFEEFCASALSVYQLEAMETWEQHARRAYELFEKDGNRPIMIEELASEL 533
Query 173 GDSYGGVLAEEILQQCDIKKNGVIDFEEFMVALTG 207
G V +LQ +G + F F+ L G
Sbjct 534 GLG-PSVPVHVVLQDWIRHSDGKLSFLGFVRLLHG 567
> Hs4885109
Length=152
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/140 (30%), Positives = 74/140 (52%), Gaps = 10/140 (7%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRID---LTGDQEINYS 130
E ++ F D+ G+I + L V+ ++L N EAE + + +D L G+ I++
Sbjct 12 EFKEAFSLFDKDGDGTITTKELGTVM-RSLGQNPTEAELQDMINEVDADDLPGNGTIDFP 70
Query 131 EFLAATF-QTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQ 186
EFL + + S+ IREAF D D +G IS LRH+ LG+ +E+++
Sbjct 71 EFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIR 130
Query 187 QCDIKKNGVIDFEEFMVALT 206
+ DI +G +++EEF+ +T
Sbjct 131 EADIDGDGQVNYEEFVQMMT 150
> At5g37780
Length=149
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 72/135 (53%), Gaps = 7/135 (5%)
Query 74 LDELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSE 131
+ E ++ F D+ G I + L V+ ++L N EAE + + +D G+ I++ E
Sbjct 10 ISEFKEAFSLFDKDGDGCITTKELGTVM-RSLGQNPTEAELQDMINEVDADGNGTIDFPE 68
Query 132 FLA-ATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQ 187
FL + + S+ ++EAF D DQ+G IS LRH+ LG+ EE++++
Sbjct 69 FLNLMAKKMKDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIRE 128
Query 188 CDIKKNGVIDFEEFM 202
D+ +G I++EEF+
Sbjct 129 ADVDGDGQINYEEFV 143
> At1g66410
Length=149
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 72/135 (53%), Gaps = 7/135 (5%)
Query 74 LDELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSE 131
+ E ++ F D+ G I + L V+ ++L N EAE + + +D G+ I++ E
Sbjct 10 ISEFKEAFSLFDKDGDGCITTKELGTVM-RSLGQNPTEAELQDMINEVDADGNGTIDFPE 68
Query 132 FLA-ATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQ 187
FL + + S+ ++EAF D DQ+G IS LRH+ LG+ EE++++
Sbjct 69 FLNLMAKKMKDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIRE 128
Query 188 CDIKKNGVIDFEEFM 202
D+ +G I++EEF+
Sbjct 129 ADVDGDGQINYEEFV 143
> CE08132
Length=218
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 39/170 (22%), Positives = 80/170 (47%), Gaps = 16/170 (9%)
Query 52 AACTAIQRAALGL----------IAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVL 101
+ C AI+ + + I + +++DE + F D+ G+I + L +
Sbjct 48 SGCKAIRAERMAIPSNLMQFSEDIIKQLTPEEIDEFREAFMMFDKDGNGTISTKEL-GIA 106
Query 102 VKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFLAATFQTQVALSQSLIREAFERLDVDQ 159
+++L N E E + + +D+ G+ +I + EF + +IREAF D D
Sbjct 107 MRSLGQNPTEQEILEMINEVDIDGNGQIEFPEFCVMMKRMMKETDSEMIREAFRVFDKDG 166
Query 160 SGEISLDNLRHI---LGDSYGGVLAEEILQQCDIKKNGVIDFEEFMVALT 206
+G I+ R+ +G + +E++++ D+ +G ID+EEF+ ++
Sbjct 167 NGVITAQEFRYFMVHMGMQFSEEEVDEMIKEVDVDGDGEIDYEEFVKMMS 216
> At1g49580
Length=606
Score = 56.2 bits (134), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 52/218 (23%), Positives = 100/218 (45%), Gaps = 18/218 (8%)
Query 1 AKDFISSLLRRNPEDRLTAAAALEHPWLAVEDGSIQRHTIDIEVLKSMRRFAACTAIQRA 60
AKDF+ LL ++P R++A+ AL HPW+ + + DI + + M+ + +++++A
Sbjct 385 AKDFVKRLLFKDPRRRMSASQALMHPWIRAYNTDMNI-PFDILIFRQMKAYLRSSSLRKA 443
Query 61 ALGLIAMSMPSKDLDELEKLFCALDEKKTGSIRVESLVNVL-------VKTLNIPEAEAH 113
AL ++ ++ ++ L+ F L K G I ++++ L +K IPE A
Sbjct 444 ALRALSKTLIKDEILYLKTQFSLLAPNKDGLITMDTIRMALASNATEAMKESRIPEFLA- 502
Query 114 RIFHRIDLTGDQEINYSEFLAATFQTQ----VALSQSLIREAFERLDVDQSGEISLDNLR 169
+ + + G +++ EF AA + + IR A+E D + + I ++ L
Sbjct 503 -LLNGLQYRG---MDFEEFCAAAINVHQHESLDCWEQSIRHAYELFDKNGNRAIVIEELA 558
Query 170 HILGDSYGGVLAEEILQQCDIKKNGVIDFEEFMVALTG 207
LG + +L +G + F F+ L G
Sbjct 559 SELGVG-PSIPVHSVLHDWIRHTDGKLSFFGFVKLLHG 595
> At3g56800
Length=149
Score = 55.8 bits (133), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 40/136 (29%), Positives = 72/136 (52%), Gaps = 7/136 (5%)
Query 76 ELEKLFCALDEKKTGSIRVESLVNVLVKTL--NIPEAEAHRIFHRIDLTGDQEINYSEFL 133
E ++ F D+ G I + L V+ ++L N EAE + + +D G+ I++ EFL
Sbjct 12 EFKEAFSLFDKDGDGCITTKELGTVM-RSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query 134 A-ATFQTQVALSQSLIREAFERLDVDQSGEISLDNLRHI---LGDSYGGVLAEEILQQCD 189
+ + S+ ++EAF D DQ+G IS LRH+ LG+ +E++++ D
Sbjct 71 NLMARKMKDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEAD 130
Query 190 IKKNGVIDFEEFMVAL 205
+ +G I++EEF+ +
Sbjct 131 VDGDGQINYEEFVKVM 146
Lambda K H
0.317 0.130 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6127321958
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40