bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0146_orf1
Length=120
Score E
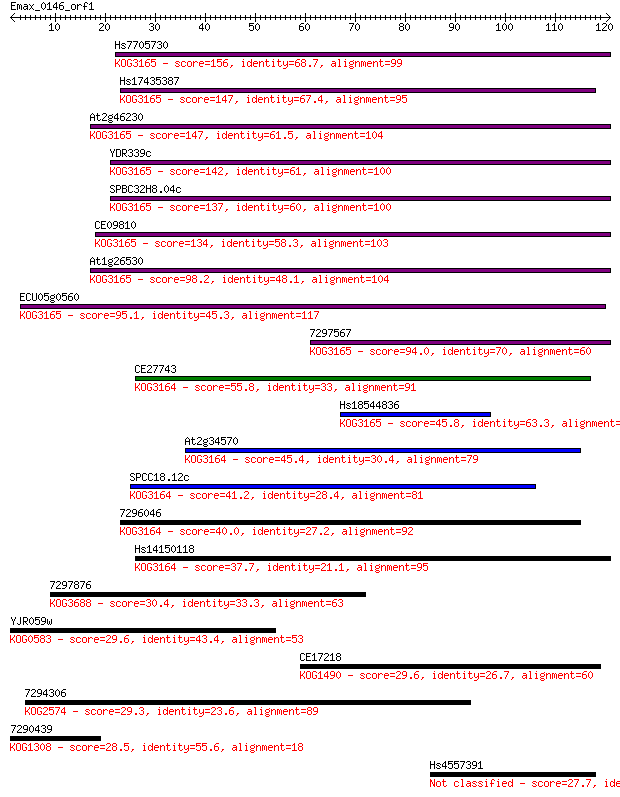
Sequences producing significant alignments: (Bits) Value
Hs7705730 156 8e-39
Hs17435387 147 4e-36
At2g46230 147 7e-36
YDR339c 142 1e-34
SPBC32H8.04c 137 4e-33
CE09810 134 5e-32
At1g26530 98.2 3e-21
ECU05g0560 95.1 3e-20
7297567 94.0 7e-20
CE27743 55.8 2e-08
Hs18544836 45.8 2e-05
At2g34570 45.4 2e-05
SPCC18.12c 41.2 5e-04
7296046 40.0 0.001
Hs14150118 37.7 0.005
7297876 30.4 0.73
YJR059w 29.6 1.3
CE17218 29.6 1.3
7294306 29.3 1.8
7290439 28.5 2.9
Hs4557391 27.7 5.0
> Hs7705730
Length=198
Score = 156 bits (395), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 68/100 (68%), Positives = 87/100 (87%), Gaps = 1/100 (1%)
Query 22 SSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQ 81
S +FF+YN QLGPP+ ++ DTNF+NFSIK KLD+ Q+MMDCL AKCIP +TDC++AE++
Sbjct 52 PSCLFFQYNTQLGPPYHILVDTNFINFSIKAKLDLVQSMMDCLYAKCIPCITDCVMAEIE 111
Query 82 KLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
KLGQKYR+AL+I K+ +FERLPC HKGTYADDC+V RV+Q
Sbjct 112 KLGQKYRVALRIAKDPRFERLPCTHKGTYADDCLVQRVTQ 151
> Hs17435387
Length=136
Score = 147 bits (371), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 64/96 (66%), Positives = 84/96 (87%), Gaps = 1/96 (1%)
Query 23 STMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQK 82
S +FF+YN QLGPP+ ++ DTNF+NFSIK KLD+ Q+MMDCL AKCIP +TDC++AE++K
Sbjct 41 SHLFFQYNTQLGPPYHILIDTNFINFSIKAKLDLMQSMMDCLYAKCIPCITDCVMAEIEK 100
Query 83 LGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNR 117
LGQKYR+AL+I K+ +FE LPCAH+GTYADDC+V +
Sbjct 101 LGQKYRVALRITKDPRFELLPCAHQGTYADDCLVQK 136
> At2g46230
Length=216
Score = 147 bits (370), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 64/105 (60%), Positives = 85/105 (80%), Gaps = 1/105 (0%)
Query 17 HAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCI 76
+ +V + +FF YN L PP+RV+ DTNF+NFSI+NK+D+ + M DCL A C P +TDC+
Sbjct 43 NVPSVPAGLFFSYNSTLVPPYRVLVDTNFINFSIQNKIDLEKGMRDCLYANCTPCITDCV 102
Query 77 IAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
+AEL+KLGQKYR+AL+I K+ FERLPC HKGTYADDC+V+RV+Q
Sbjct 103 MAELEKLGQKYRVALRIAKDPHFERLPCIHKGTYADDCLVDRVTQ 147
> YDR339c
Length=189
Score = 142 bits (358), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 61/101 (60%), Positives = 86/101 (85%), Gaps = 1/101 (0%)
Query 21 VSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAEL 80
VSS +FF+YN + PP++V+ DTNF+NFSI+ K+D+ + MMDCLLAKC P++TDC++AEL
Sbjct 47 VSSALFFQYNQAIKPPYQVLIDTNFINFSIQKKVDIVRGMMDCLLAKCNPLITDCVMAEL 106
Query 81 QKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
+KLG KYR+ALK+ ++ + +RL C+HKGTYADDC+V+RV Q
Sbjct 107 EKLGPKYRIALKLARDPRIKRLSCSHKGTYADDCLVHRVLQ 147
> SPBC32H8.04c
Length=192
Score = 137 bits (346), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 60/101 (59%), Positives = 82/101 (81%), Gaps = 1/101 (0%)
Query 21 VSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAEL 80
++S +FF++N LGPP+ VI DTNF+NF ++ K+D+ + +M CL AK IP ++DC++AEL
Sbjct 49 MASNLFFQFNESLGPPYHVIIDTNFINFCLQQKIDLFEGLMTCLYAKTIPCISDCVMAEL 108
Query 81 QKLGQKYRLALKIVK-EQFERLPCAHKGTYADDCIVNRVSQ 120
+KLG +YR+AL+I K E+FERLPC HKGTYADDCIV RV Q
Sbjct 109 EKLGIRYRIALRIAKDERFERLPCTHKGTYADDCIVQRVMQ 149
> CE09810
Length=196
Score = 134 bits (336), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 60/104 (57%), Positives = 83/104 (79%), Gaps = 2/104 (1%)
Query 18 AATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCII 77
A +SS MF +YN QLGPPF VI DTNFVNF++KN++D+ Q MDCL AK I TDC++
Sbjct 47 APKISSAMFMKYNTQLGPPFHVIVDTNFVNFAVKNRIDMFQGFMDCLFAKTIVYATDCVL 106
Query 78 AELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
AEL+K+ +++++ALK++K+ + +RL C HKGTYADDC+V RV+Q
Sbjct 107 AELEKV-RRFKIALKVLKDPRVQRLKCEHKGTYADDCLVQRVTQ 149
> At1g26530
Length=189
Score = 98.2 bits (243), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 50/105 (47%), Positives = 66/105 (62%), Gaps = 27/105 (25%)
Query 17 HAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCI 76
+ +V S +FF +N L PP+RV+ DTNF+NFSI+NK
Sbjct 43 NVPSVPSGLFFSHNSSLVPPYRVLVDTNFINFSIQNK----------------------- 79
Query 77 IAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVSQ 120
L+KLGQKYR+AL+I K+ +FERLPC KGTYADDC+V+RV+Q
Sbjct 80 ---LEKLGQKYRVALRIAKDPRFERLPCVLKGTYADDCLVDRVTQ 121
> ECU05g0560
Length=179
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 53/122 (43%), Positives = 73/122 (59%), Gaps = 8/122 (6%)
Query 3 EREKERAKK-EQAKHHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMM 61
+R+K R K E A +T F N L PP+ +I DTNF+N I+ K D+ + +M
Sbjct 17 DRKKTRVKSPENASSTGENPRNTEDFLLNHTLRPPYNIILDTNFINDCIRKKYDIREQLM 76
Query 62 DCLLAK---CIPMVTDCIIAELQKLGQKYRLALKIVKEQ-FERLPCAHKGTYADDCIVNR 117
+ A CIP DC+I EL+ LG+ +R+AL +++ +RL C HKGTYADDCI NR
Sbjct 77 KAVNASIKICIP---DCVIGELESLGRPFRVALASIRDSDVQRLECDHKGTYADDCIFNR 133
Query 118 VS 119
VS
Sbjct 134 VS 135
> 7297567
Length=107
Score = 94.0 bits (232), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 42/61 (68%), Positives = 52/61 (85%), Gaps = 1/61 (1%)
Query 61 MDCLLAKCIPMVTDCIIAELQKLGQKYRLALKIVKE-QFERLPCAHKGTYADDCIVNRVS 119
MDCL AKCIP ++DC+ AEL+KLG KY+LAL+I+ + +FERLPC HKGTYADDC+V RV
Sbjct 1 MDCLYAKCIPYISDCVRAELEKLGNKYKLALRIISDPRFERLPCLHKGTYADDCLVERVR 60
Query 120 Q 120
Q
Sbjct 61 Q 61
> CE27743
Length=232
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 49/92 (53%), Gaps = 2/92 (2%)
Query 26 FFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQ 85
F++YN + P+RV+ D F N +++ KL++ + + L + M T+C++ EL+K G
Sbjct 15 FYKYNYKFVAPYRVLVDGTFCNAALQEKLNLAEQIPKYLTEETHLMTTNCVLKELEKFGP 74
Query 86 KYRLALKIVKEQFERLPCAHKGT-YADDCIVN 116
A I K QFE C H A DC+ +
Sbjct 75 LLYGAFVIAK-QFEIAECTHSTPRAASDCLAH 105
> Hs18544836
Length=141
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 19/30 (63%), Positives = 26/30 (86%), Gaps = 0/30 (0%)
Query 67 KCIPMVTDCIIAELQKLGQKYRLALKIVKE 96
KCIP +TDC++AE+ KLGQKY A++IVK+
Sbjct 108 KCIPCITDCVMAEIGKLGQKYGAAVRIVKD 137
> At2g34570
Length=258
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 36 PFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPM-VTDCIIAELQKLGQKYRLALKIV 94
P++V+CD FV+ + N++ + LL + + T C+IAEL+KLG+ + +L+
Sbjct 25 PYKVLCDGTFVHHLVTNEITPADTAVSELLGGPVKLFTTRCVIAELEKLGKDFAESLEAA 84
Query 95 KEQFERLPCAH-KGTYADDCI 114
+ C H + AD+C+
Sbjct 85 -QTLNTATCEHEEAKTADECL 104
> SPCC18.12c
Length=260
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 40/83 (48%), Gaps = 3/83 (3%)
Query 25 MFFRYNLQLG--PPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQK 82
+ Y L G P++V+ D +F+ + K+D+ A+ + PM+T C I +L
Sbjct 12 LMHTYQLLFGFREPYQVLVDADFLKDLSQQKIDIQAALARTVQGAIKPMITQCCIRQLYS 71
Query 83 LGQKYRLALKIVKEQFERLPCAH 105
+ + ++I K FER C H
Sbjct 72 KSDELKQEIRIAKS-FERRRCGH 93
> 7296046
Length=244
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 43/94 (45%), Gaps = 3/94 (3%)
Query 23 STMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQK 82
+ +FF N P++V+ D F +++ K+ + + + + T C+I E +
Sbjct 12 TLVFFASNFDYREPYQVLIDATFCQAALQQKIGIDEQIKKYFQCGVKLLTTQCVILESES 71
Query 83 LGQKYRLALKIVKEQFERLPCAHKG--TYADDCI 114
LG A IVK +F C H+G A +CI
Sbjct 72 LGAPLTGATSIVK-RFHVHKCGHEGKPVPASECI 104
> Hs14150118
Length=249
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/97 (20%), Positives = 49/97 (50%), Gaps = 3/97 (3%)
Query 26 FFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMMDCLLAKCIPMVTDCIIAELQKLGQ 85
FFR N + P++++ D F +++ ++ + + + L+ + T C++ EL+ LG+
Sbjct 15 FFRNNFGVREPYQILLDGTFCQAALRGRIQLREQLPRYLMGETQLCTTRCVLKELETLGK 74
Query 86 KYRLALKIVKEQFERLPCAH--KGTYADDCIVNRVSQ 120
A K++ ++ + C H +C+++ V +
Sbjct 75 DLYGA-KLIAQKCQVRNCPHFKNAVSGSECLLSMVEE 110
> 7297876
Length=605
Score = 30.4 bits (67), Expect = 0.73, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 33/76 (43%), Gaps = 13/76 (17%)
Query 9 AKKEQAKHHAATVSSTMFFR---YNLQLGPPFRVICDTN----------FVNFSIKNKLD 55
AK+ H + FFR +LG PF +CD N F++F ++ +
Sbjct 393 AKQWGVHHRWTMLLLEEFFRQGDLEKELGLPFSPLCDRNNTLVAESQICFIDFIVEPSMG 452
Query 56 VCQAMMDCLLAKCIPM 71
V M++ +LA PM
Sbjct 453 VMSDMLELILAPIAPM 468
> YJR059w
Length=818
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 1 DREREKERAK-KEQAKHHAA------TVSSTMFFRYNLQLGP----PFRVICDTNFVNFS 49
DRE+EKERA+ KE+ HHA +++S F N+ P P DT F +
Sbjct 145 DREKEKERARNKERNTHHAGLPQRSNSMASHHFPNENIVYNPYGISPNHARPDTAFADTL 204
Query 50 IKNK 53
NK
Sbjct 205 NTNK 208
> CE17218
Length=681
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 59 AMMDCLLAKCIPMVTDCIIAELQKLGQKYRLALKIVKEQFERLPCAHKGTYADDCIVNRV 118
A++D L + IP++ + + +G + R +++ ++ E A K T +DC++NRV
Sbjct 306 ALLDQLEKEGIPIIETSTLTQEGVMGLRDRACDELLAQRVEAKIQAKKITNVEDCVLNRV 365
> 7294306
Length=501
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 21/91 (23%), Positives = 41/91 (45%), Gaps = 7/91 (7%)
Query 4 REKERAKK--EQAKHHAATVSSTMFFRYNLQLGPPFRVICDTNFVNFSIKNKLDVCQAMM 61
R+ ER K +Q +H+A+ + +++ P + +I D N + I N++ +
Sbjct 63 RDSERLKNTLQQIEHYASRQRTAAEMLGSVESDPEYCLIVDANAIAVDIDNEISIVHKFT 122
Query 62 DCLLAKCIPMVTDCIIAELQKLGQKYRLALK 92
K P + I+ E++ Y LA+K
Sbjct 123 KEKYQKRFPELDSLIVGEIE-----YLLAVK 148
> 7290439
Length=373
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 10/18 (55%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 1 DREREKERAKKEQAKHHA 18
+R+R++ RA+KEQ KH+A
Sbjct 257 ERQRDQRRARKEQEKHNA 274
> Hs4557391
Length=591
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 16/33 (48%), Gaps = 0/33 (0%)
Query 85 QKYRLALKIVKEQFERLPCAHKGTYADDCIVNR 117
++YR A + QF PC +DC+ NR
Sbjct 82 KRYRYAYLLQPSQFHGEPCNFSDKEVEDCVTNR 114
Lambda K H
0.326 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40