bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0155_orf3
Length=254
Score E
Sequences producing significant alignments: (Bits) Value
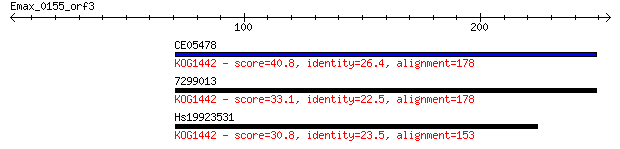
CE05478 40.8 0.003
7299013 33.1 0.51
Hs19923531 30.8 3.1
> CE05478
Length=416
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 47/183 (25%), Positives = 82/183 (44%), Gaps = 10/183 (5%)
Query 71 PVFVTWWQLAQGLLMAWCLGEVGKEYPKLAYFPRVEIDRRLLRRLFVPTIVNALMLVLAN 130
P+F+TW+Q + + L + K Y L FP + ID ++ R + ++V M+ N
Sbjct 113 PLFITWYQCLVTVFLCLFLSKTSKAYG-LFKFPSMPIDAKISREVLPLSVVFVAMISFNN 171
Query 131 VLLYRVQCVATLPVVVAFAVVLHHVTRFVGCGEEYMPMRWQAVG---LLFTAFLLGITDS 187
+ L V V+ V + V + V ++ G++ QA+G L+ FLLG+
Sbjct 172 LCLKYVG-VSFYYVGRSLTTVFNVVCTYLILGQK---TSGQAIGCCALIIFGFLLGVDQE 227
Query 188 KTVGSDVLPWAI--LYAIFSAAFRAAFLQKVMHEVEGKGNLLHNHQHFISVIILPVLMLI 245
G+ I + A S A A + +KV+ V L + + ++++ LML
Sbjct 228 GVTGTLSYTGVIFGVLASLSVALNAIYTRKVLSSVGDCLWRLTMYNNLNALVLFLPLMLF 287
Query 246 CGE 248
GE
Sbjct 288 NGE 290
> 7299013
Length=337
Score = 33.1 bits (74), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 40/190 (21%), Positives = 84/190 (44%), Gaps = 22/190 (11%)
Query 71 PVFVTWWQLAQGLLMAWCLGEVGKEYPKLAYFPRVE-IDRRLLRRLFVPTIVNALMLVLA 129
P+F++W+Q ++ + + ++YP + FP +D R++ +++ LM+
Sbjct 50 PLFMSWFQCVVSTVICFVASRLSRKYPSVFTFPEGNPLDIDTFRKILPLSVLYTLMIGAN 109
Query 130 NVLLYRVQCVATLPVVVAFAVVLHHVTRFVGCGEEYMPMRWQA-------VGLLFTAFLL 182
N+ L V VAF + +T Y+ +R + G + F L
Sbjct 110 NLSLSY--------VTVAFYYIGRSLTTVFSVVLTYVILRQRTSFKCLLCCGAIVVGFWL 161
Query 183 GITDSKTVGSDVLPW-AILYAIFSAAFRAAF---LQKVMHEVEGKGNLLHNHQHFISVII 238
G+ D +++ ++V W ++ + S+ A F +K + V + LL + + S ++
Sbjct 162 GV-DQESL-TEVFSWRGTIFGVLSSLALAMFSIQTKKSLGYVNQEVWLLSYYNNLYSTLL 219
Query 239 LPVLMLICGE 248
L++I GE
Sbjct 220 FLPLIIINGE 229
> Hs19923531
Length=351
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 36/164 (21%), Positives = 66/164 (40%), Gaps = 21/164 (12%)
Query 71 PVFVTWWQLAQGLLMAWCLGEVGKEYPKLAYFPRVEIDRRLLRRLFVPTIVNALMLVLAN 130
P+FVT++Q L+ L + P FP + +D R+ R + ++V M+ N
Sbjct 60 PIFVTFYQCLVTTLLCKGLSALAACCPGAVDFPSLRLDLRVARSVLPLSVVFIGMITFNN 119
Query 131 VLLYRVQCVATLPVVVAFAVVLHHVTRFVGCGEEYMPMRWQ-------AVGLLFTAFLLG 183
+ L V VAF V +T Y+ ++ G++ F LG
Sbjct 120 LCLKYVG--------VAFYNVGRSLTTVFNVLLSYLLLKQTTSFYALLTCGIIIGGFWLG 171
Query 184 ITDSKTVGSDVLPW-AILYAIFSA---AFRAAFLQKVMHEVEGK 223
+ G+ L W ++ + ++ + A + KV+ V+G
Sbjct 172 VDQEGAEGT--LSWLGTVFGVLASLCVSLNAIYTTKVLPAVDGS 213
Lambda K H
0.331 0.143 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5258582968
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40