bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0169_orf1
Length=247
Score E
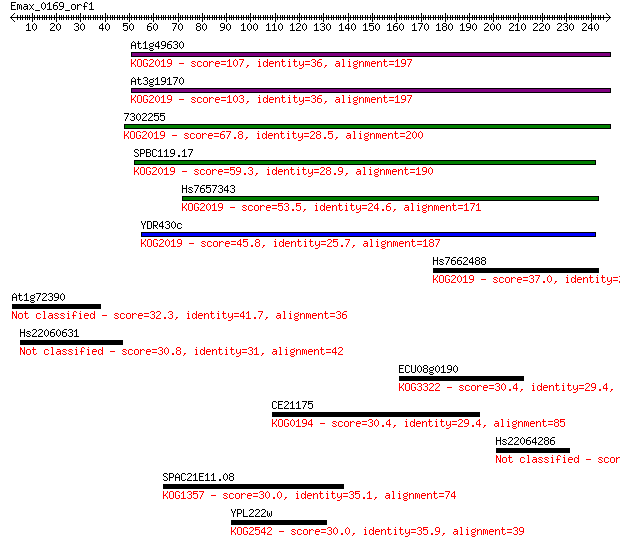
Sequences producing significant alignments: (Bits) Value
At1g49630 107 2e-23
At3g19170 103 3e-22
7302255 67.8 2e-11
SPBC119.17 59.3 7e-09
Hs7657343 53.5 3e-07
YDR430c 45.8 8e-05
Hs7662488 37.0 0.039
At1g72390 32.3 0.88
Hs22060631 30.8 2.6
ECU08g0190 30.4 3.3
CE21175 30.4 3.6
Hs22064286 30.0 4.6
SPAC21E11.08 30.0 4.9
YPL222w 30.0 5.2
> At1g49630
Length=1076
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 71/199 (35%), Positives = 115/199 (57%), Gaps = 5/199 (2%)
Query 51 LNGEERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVP 110
L+ + +L L L+ L++G ASPL L + LG G++ +L F++GLK V
Sbjct 391 LDLQTQLALGFLDHLMLGTPASPLRKILLE-SGLGEALVNSGMEDELLQPQFSIGLKGV- 448
Query 111 QREGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMT 170
+ ++VEELV+NTL+++ ++GF + ++ A++N IEF LRE PRGL +
Sbjct 449 -SDDNVQKVEELVMNTLRKLADEGFDTDAVEASMNTIEFSLRENNTGSSPRGLSLMLQSI 507
Query 171 QEIVYNKDPLNALEFEETLWKIRERLA-KGEK-VFEQLIQKYFSDNPHRLTLRLIGNKEI 228
+ +Y+ DP L++EE L ++ R+A KG K VF LI++Y +NPH +T+ + + E
Sbjct 508 AKWIYDMDPFEPLKYEEPLKSLKARIAEKGSKSVFSPLIEEYILNNPHCVTIEMQPDPEK 567
Query 229 IKEREEEEKKSLKKAEKEM 247
E EEK L+K + M
Sbjct 568 ASLEEAEEKSILEKVKASM 586
> At3g19170
Length=1052
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 71/199 (35%), Positives = 113/199 (56%), Gaps = 5/199 (2%)
Query 51 LNGEERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVP 110
L+ + +L L L+ L++G ASPL L + LG + GL +L F +GLK V
Sbjct 368 LDLQTQLALGFLDHLMLGTPASPLRKILLE-SGLGEALVSSGLSDELLQPQFGIGLKGV- 425
Query 111 QREGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMT 170
E ++VEEL+++TLK++ E+GF + ++ A++N IEF LRE PRGL +
Sbjct 426 -SEENVQKVEELIMDTLKKLAEEGFDNDAVEASMNTIEFSLRENNTGSFPRGLSLMLQSI 484
Query 171 QEIVYNKDPLNALEFEETLWKIRERLA-KGEK-VFEQLIQKYFSDNPHRLTLRLIGNKEI 228
+ +Y+ DP L++ E L ++ R+A +G K VF LI+K +N HR+T+ + + E
Sbjct 485 SKWIYDMDPFEPLKYTEPLKALKTRIAEEGSKAVFSPLIEKLILNNSHRVTIEMQPDPEK 544
Query 229 IKEREEEEKKSLKKAEKEM 247
+ E EEK L+K + M
Sbjct 545 ATQEEVEEKNILEKVKAAM 563
> 7302255
Length=1112
Score = 67.8 bits (164), Expect = 2e-11, Method: Composition-based stats.
Identities = 57/200 (28%), Positives = 93/200 (46%), Gaps = 13/200 (6%)
Query 48 CTLLNGEERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLK 107
C N +E L VL+ +LI SP Y +L GG T G D K F VGL+
Sbjct 360 CDATNIQESFELHVLSEVLIRGPNSPFYKNLIEPNFSGGYNQTTGYSSDTKDTTFVVGLQ 419
Query 108 DVPQREGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFAR 167
D+ R K+ E+ T+ GF S + + L+ +E L+ L+F
Sbjct 420 DL--RVEDFKKCIEIFDKTIINSMNDGFDSQHVESVLHNLELSLKHQ-NPNFGNTLLF-- 474
Query 168 KMTQEIVYNKDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKE 227
T ++ D ++ L + + +RE +++ +K F++ I+KYF++N HRLTL +
Sbjct 475 NSTALWNHDGDVVSNLRVSDMISGLRESISQNKKYFQEKIEKYFANNNHRLTLTM----- 529
Query 228 IIKEREEEEKKSLKKAEKEM 247
+E + K+AE E+
Sbjct 530 ---SPDEAYEDKFKQAELEL 546
> SPBC119.17
Length=882
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 55/201 (27%), Positives = 97/201 (48%), Gaps = 28/201 (13%)
Query 52 NGEERLLLQVLNFLLIGDKASPLYSSLSNYTQLG-GPAYTP--GLDLDLKYAMFTVGLKD 108
N E L+VL+ L +SP Y +L + G G + P G D K +F+VGL
Sbjct 324 NVYETFALKVLSKLCFDGFSSPFYKAL---IESGLGTDFAPNSGYDSTTKRGIFSVGL-- 378
Query 109 VPQREGAAKE----VEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLI 164
EGA++E +E LV + ++ KGF + L A L+++E L+ +
Sbjct 379 ----EGASEESLAKIENLVYSIFNDLALKGFENEKLEAILHQMEISLKH-------KSAH 427
Query 165 FARKMTQEIVYN----KDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTL 220
F + Q + +N DP + L F + + ++++ + G K+F++LI+KY +N R
Sbjct 428 FGIGLAQSLPFNWFNGADPADWLSFNKQIEWLKQKNSDG-KLFQKLIKKYILENKSRFVF 486
Query 221 RLIGNKEIIKEREEEEKKSLK 241
++ + + +E E K L+
Sbjct 487 TMLPSSTFPQRLQEAEAKKLQ 507
> Hs7657343
Length=1038
Score = 53.5 bits (127), Expect = 3e-07, Method: Composition-based stats.
Identities = 42/173 (24%), Positives = 84/173 (48%), Gaps = 9/173 (5%)
Query 72 SPLYSSLSNYTQLGGPAYTP--GLDLDLKYAMFTVGLKDVPQREGAAKEVEELVINTLKE 129
SP Y +L G ++P G + + A F+VGL+ + +++ + V L+ T+ E
Sbjct 359 SPFYKALIESGL--GTEFSPDVGYNGYTREAYFSVGLQGIVEKD--IETVRSLIDRTIDE 414
Query 130 IKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDPLNALEFEETL 189
+ E + A L+KIE ++ GL+ + ++ DP+ L+ L
Sbjct 415 VVETRIEDDRIEALLHKIEIQMKHQSTSF---GLMLTSYIASCWNHDGDPVELLKLGNQL 471
Query 190 WKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKEREEEEKKSLKK 242
K R+ L + K ++ +++YF +N H+LTL + + + +++ + E LK+
Sbjct 472 AKFRQCLQENPKFLQEKVKQYFKNNQHKLTLSMRPDDKYHEKQAQVEATKLKQ 524
> YDR430c
Length=989
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 48/188 (25%), Positives = 82/188 (43%), Gaps = 7/188 (3%)
Query 55 ERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQREG 114
+ LL+VL LL+ +S +Y L + G++ + TVG++ V E
Sbjct 317 DTFLLKVLGNLLMDGHSSVMYQKLIESGIGLEFSVNSGVEPTTAVNLLTVGIQGVSDIEI 376
Query 115 AAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIV 174
V + N L+ E F + A + ++E ++ D GL +
Sbjct 377 FKDTVNNIFQNLLE--TEHPFDRKRIDAIIEQLELSKKDQKADF---GLQLLYSILPGWT 431
Query 175 YNKDPLNALEFEETLWKIRERL-AKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKERE 233
DP +L FE+ L + R L KG+ +F+ LI+KY P T + G++E K +
Sbjct 432 NKIDPFESLLFEDVLQRFRGDLETKGDTLFQDLIRKYIVHKP-CFTFSIQGSEEFSKSLD 490
Query 234 EEEKKSLK 241
+EE+ L+
Sbjct 491 DEEQTRLR 498
> Hs7662488
Length=595
Score = 37.0 bits (84), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 39/68 (57%), Gaps = 0/68 (0%)
Query 175 YNKDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKEREE 234
++ DP+ L+ L K R+ L + K ++ +++YF +N H+LTL + + + +++ +
Sbjct 15 HDGDPVELLKLGNQLAKFRQCLQENPKFLQEKVKQYFKNNQHKLTLSMRPDDKYHEKQAQ 74
Query 235 EEKKSLKK 242
E LK+
Sbjct 75 VEATKLKQ 82
> At1g72390
Length=1088
Score = 32.3 bits (72), Expect = 0.88, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 2 EYRYPSSAEELKDWVTVSWVLNPGKKEEKEGKKEEK 37
+ R P +A ++ SW +NPG++ EKE KKEE+
Sbjct 262 QSRMPHNAFIRSNFPQTSWNVNPGQQIEKEPKKEEQ 297
> Hs22060631
Length=817
Score = 30.8 bits (68), Expect = 2.6, Method: Composition-based stats.
Identities = 13/50 (26%), Positives = 27/50 (54%), Gaps = 8/50 (16%)
Query 5 YPSSAEELKDWVTVS--------WVLNPGKKEEKEGKKEEKKDSSPPSSS 46
+P ++E++ + +S W+L+P ++ G + + +DS PPS S
Sbjct 333 HPRGSQEMRKVLALSLNKHQVYLWILDPRRRSWNPGPRPDLRDSGPPSRS 382
> ECU08g0190
Length=405
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 161 RGLIFARKMTQEIVYNKDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYF 211
RG IF +VYN+ L + +L + +++ G VFE ++ K F
Sbjct 106 RGFIFDESYKDVVVYNRPNTEILGIDFSLENVVQKIKHGNIVFEAIVGKRF 156
> CE21175
Length=728
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 36/85 (42%), Gaps = 14/85 (16%)
Query 109 VPQREGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARK 168
VP R A + VE+++ + KE FS L E PR+R+P L
Sbjct 178 VPVRYMAPEAVEKMI--STKESDVYAFSVLIFEVFHKAYEPFGEYGPRERVPEAL----- 230
Query 169 MTQEIVYNKDPLNALEFEETLWKIR 193
KD + L+FEE L++ R
Sbjct 231 -------GKDEVRLLQFEEILYQFR 248
> Hs22064286
Length=415
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 201 KVFEQLIQKYFSDNPHRLTLRLIGNKEIIK 230
K F L + +DNP L LRL+G K++I+
Sbjct 385 KSFPDLARSQLTDNPAILVLRLMGGKDVIR 414
> SPAC21E11.08
Length=272
Score = 30.0 bits (66), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 35/81 (43%), Gaps = 7/81 (8%)
Query 64 FLLIGDKASPLYSSLSNY---TQLGGPAYTPGLDLDLKYAMFTVGLKDVPQREGAAKEVE 120
F L GD + L S NY Q GP T + KY + T + G KEVE
Sbjct 180 FKLTGDTSLALNVSSYNYLGFAQSHGPCATKVEEAMQKYGLSTCSSNAICGTYGLHKEVE 239
Query 121 ELVINTLKE----IKEKGFSS 137
EL N + + + +GFS+
Sbjct 240 ELTANFVGKPAALVFSQGFST 260
> YPL222w
Length=688
Score = 30.0 bits (66), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 92 GLDLDLKYAMFTVGLKDVPQREGAAKEVEELVINTLKEI 130
G+DLDL+ M + LKD+ AKE ++++ L +I
Sbjct 455 GVDLDLEKCMSSTNLKDIEHAAEKAKEFCDVIVEPLLDI 493
Lambda K H
0.311 0.132 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5009866206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40