bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0263_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
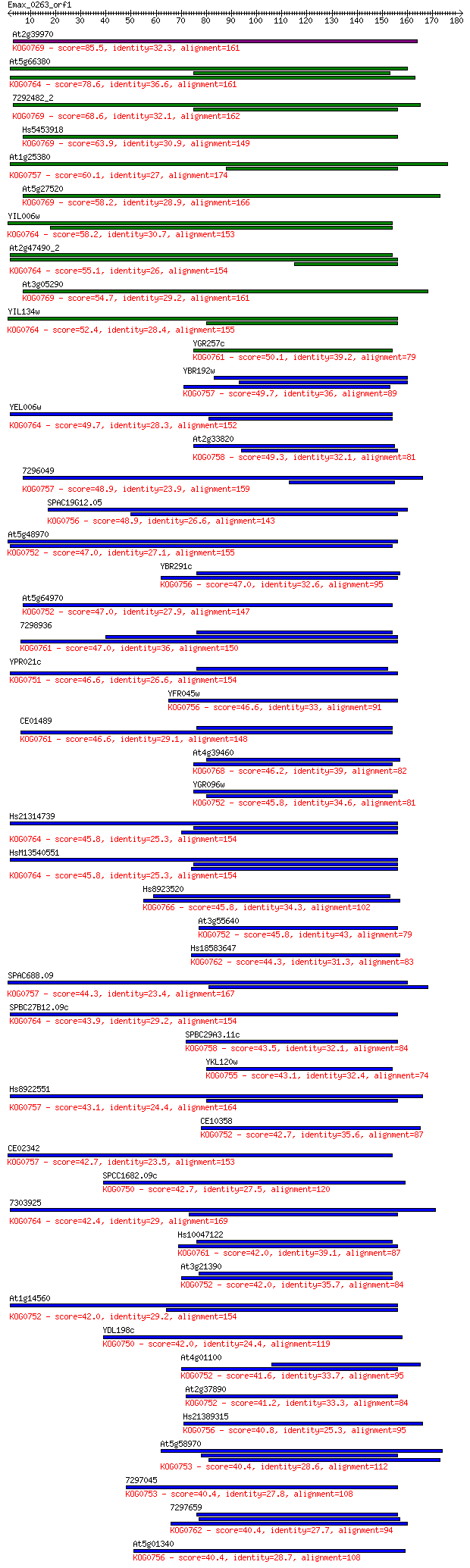
At2g39970 85.5 6e-17
At5g66380 78.6 6e-15
7292482_2 68.6 7e-12
Hs5453918 63.9 2e-10
At1g25380 60.1 2e-09
At5g27520 58.2 8e-09
YIL006w 58.2 1e-08
At2g47490_2 55.1 7e-08
At3g05290 54.7 9e-08
YIL134w 52.4 5e-07
YGR257c 50.1 2e-06
YBR192w 49.7 3e-06
YEL006w 49.7 4e-06
At2g33820 49.3 4e-06
7296049 48.9 6e-06
SPAC19G12.05 48.9 6e-06
At5g48970 47.0 2e-05
YBR291c 47.0 2e-05
At5g64970 47.0 2e-05
7298936 47.0 2e-05
YPR021c 46.6 3e-05
YFR045w 46.6 3e-05
CE01489 46.6 3e-05
At4g39460 46.2 4e-05
YGR096w 45.8 4e-05
Hs21314739 45.8 4e-05
HsM13540551 45.8 4e-05
Hs8923520 45.8 5e-05
At3g55640 45.8 5e-05
Hs18583647 44.3 1e-04
SPAC688.09 44.3 1e-04
SPBC27B12.09c 43.9 2e-04
SPBC29A3.11c 43.5 2e-04
YKL120w 43.1 3e-04
Hs8922551 43.1 3e-04
CE10358 42.7 4e-04
CE02342 42.7 4e-04
SPCC1682.09c 42.7 4e-04
7303925 42.4 5e-04
Hs10047122 42.0 6e-04
At3g21390 42.0 6e-04
At1g14560 42.0 7e-04
YDL198c 42.0 7e-04
At4g01100 41.6 9e-04
At2g37890 41.2 0.001
Hs21389315 40.8 0.002
At5g58970 40.4 0.002
7297045 40.4 0.002
7297659 40.4 0.002
At5g01340 40.4 0.002
> At2g39970
Length=331
Score = 85.5 bits (210), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 52/165 (31%), Positives = 90/165 (54%), Gaps = 31/165 (18%)
Query 3 GWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G+ G++P L++V NP++QF LY+ + KL K+R + + S+N
Sbjct 188 GFWKGVIPTLIMVSNPSMQFMLYETMLTKLK--KKR----ALKGSNN------------- 228
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKD----YSSSVRSL 118
VT E+FL+G AKL A + TYP LVVK+R Q+K D Y ++ ++
Sbjct 229 --------VTALETFLLGAVAKLGATVTTYPLLVVKSRLQAKQVTTGDKRQQYKGTLDAI 280
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLF 163
+ ++ EGL G + G+ +K+ ++L++A++F + E+L+ LL
Sbjct 281 LKMIRYEGLYGFYKGMSTKIVQSVLAAAVLFMIKEELVKGAKLLL 325
> At5g66380
Length=310
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 57/161 (35%), Positives = 85/161 (52%), Gaps = 26/161 (16%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
R GIVP LVLV + AIQFT Y+ LR ++ LK+R R + E + N L+S
Sbjct 165 RALYKGIVPGLVLVSHGAIQFTAYEELRKIIVDLKERRRKS--ESTDNLLNS-------- 214
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
A+ +G ++K+AA L+TYP+ V++ R Q + + + SL +I
Sbjct 215 ------------ADYAALGGSSKVAAVLLTYPFQVIRARLQQRP-STNGIPRYIDSLHVI 261
Query 122 LET---EGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLV 159
ET EGL G + GL + L + +S+I F VYE +L L+
Sbjct 262 RETARYEGLRGFYRGLTANLLKNVPASSITFIVYENVLKLL 302
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 36/81 (44%), Gaps = 3/81 (3%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQ---SKVYNVKDYSSSVRSLIMILETEGLSGLF 131
E+ G A A + VV+TR Q + ++ Y ++ ++ I EGL GL+
Sbjct 8 ENATAGAVAGFATVAAMHSLDVVRTRFQVNDGRGSSLPTYKNTAHAVFTIARLEGLRGLY 67
Query 132 AGLPSKLTATLLSSAIMFTVY 152
AG + + +S + F Y
Sbjct 68 AGFFPAVIGSTVSWGLYFFFY 88
Score = 30.0 bits (66), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 45/166 (27%), Positives = 69/166 (41%), Gaps = 36/166 (21%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG AG PA V+ + + LY + Q R R + + L+S
Sbjct 64 RGLYAGFFPA---VIGSTVSWGLYFFFYGRAKQRYARGRDDEKLSPALHLAS-------- 112
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSS----VRS 117
AE+ +G+ L+ + YL Q+KV D SSS +R+
Sbjct 113 -----------AAEAGALGMM--LSGLFMHKSYLAC----QNKV-TASDTSSSNSTILRA 154
Query 118 LIMILETEGLSGLFAGLPSKLTATLLS-SAIMFTVYEQLLPLVVLL 162
+ I++ EG L+ G+ L L+S AI FT YE+L ++V L
Sbjct 155 IRTIVKEEGPRALYKGIVPGLV--LVSHGAIQFTAYEELRKIIVDL 198
> 7292482_2
Length=301
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 52/173 (30%), Positives = 85/173 (49%), Gaps = 42/173 (24%)
Query 3 GWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G +G +P+L+LV NPA+QF +Y+ L+ +M R E S S
Sbjct 158 GLWSGTIPSLMLVSNPALQFMMYEMLKRNIM------RFTGGEMGSLSF----------- 200
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT--RAQSKVYNVKDYSSSVRS--- 117
F +G AK A ++TYP +V+T R +SK + K +S+ +
Sbjct 201 --------------FFIGAIAKAFATVLTYPLQLVQTKQRHRSKESDSKPSTSAGSTPRT 246
Query 118 ------LIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFR 164
+I IL+ +G+ GLF GL +K+ T+L++A+MF YE++ V +L +
Sbjct 247 ESTLELMISILQHQGIRGLFRGLEAKILQTVLTAALMFMAYEKIAGTVGMLLK 299
Score = 35.4 bits (80), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 47/90 (52%), Gaps = 13/90 (14%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNV--------KDYSSSVRSLIMILETEG 126
+ L+G A + L T P+ VV TR ++ NV K Y + + L + E EG
Sbjct 98 KDLLLGSIAGIINVLTTTPFWVVNTRL--RMRNVAGTSDEVNKHYKNLLEGLKYVAEKEG 155
Query 127 LSGLFAG-LPSKLTATLLSSAIMFTVYEQL 155
++GL++G +PS + + + A+ F +YE L
Sbjct 156 IAGLWSGTIPSLMLVS--NPALQFMMYEML 183
> Hs5453918
Length=307
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 78/156 (50%), Gaps = 39/156 (25%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G P+L+LV NPAIQF Y+ L+ +L++
Sbjct 168 GTFPSLLLVFNPAIQFMFYEGLKRQLLKK------------------------------- 196
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT--RAQSKVYNVKDYS-SSVRSLIMILE 123
R++++ + F++G AK A VTYP V++ R N ++ + S+R+++ +L
Sbjct 197 -RMKLSSLDVFIIGAVAKAIATTVTYPLQTVQSILRFGRHRLNPENRTLGSLRNILYLLH 255
Query 124 TE----GLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
G+ GL+ GL +KL T+L++A+MF VYE+L
Sbjct 256 QRVRRFGIMGLYKGLEAKLLQTVLTAALMFLVYEKL 291
> At1g25380
Length=376
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 47/177 (26%), Positives = 84/177 (47%), Gaps = 35/177 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG +GI+P+L V + AIQF Y+ ++ + ++ ++ S ++ S G+
Sbjct 187 RGLYSGILPSLAGVSHVAIQFPAYEKIKQYMAKM-------------DNTSVENLSPGN- 232
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTR--AQSKVYNVK-DYSSSVRSL 118
V A S AK+ A+++TYP+ V++ + Q ++ N + YS + +
Sbjct 233 ---------VAIASSI-----AKVIASILTYPHEVIRAKLQEQGQIRNAETKYSGVIDCI 278
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLRVPTYRKRD 175
+ +EG+ GL+ G + L T S+ I FT YE +L FR + P + D
Sbjct 279 TKVFRSEGIPGLYRGCATNLLRTTPSAVITFTTYEMML----RFFRQVVPPETNRSD 331
Score = 32.7 bits (73), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 36/71 (50%), Gaps = 5/71 (7%)
Query 88 ALVTYPYLVVKTR--AQSKVYNVKDYSSSVRSLIMILETEGLSGLFAG-LPSKLTATLLS 144
++ T P VVKTR Q V Y S + + I EG+ GL++G LPS A +
Sbjct 145 SIATNPLWVVKTRLMTQGIRPGVVPYKSVMSAFSRICHEEGVRGLYSGILPS--LAGVSH 202
Query 145 SAIMFTVYEQL 155
AI F YE++
Sbjct 203 VAIQFPAYEKI 213
> At5g27520
Length=321
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 48/178 (26%), Positives = 86/178 (48%), Gaps = 34/178 (19%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G+ +L+L NPAIQ+T++D L+ L++ + + S + SS
Sbjct 162 GLGISLLLTSNPAIQYTVFDQLKQNLLE---KGKAKSNKDSSP----------------- 201
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ----SKVYNVKDYSSSVRSLI--- 119
+ ++ +F++G +K AA ++TYP + K Q SK K +R I
Sbjct 202 --VVLSAFMAFVLGAVSKSAATVITYPAIRCKVMIQAADDSKENEAKKPRKRIRKTIPGV 259
Query 120 --MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLP---LVVLLFRSLRVPTYR 172
I + EG+ G F GL +++ T+LSSA++ + E++ +++L R+L V R
Sbjct 260 VYAIWKKEGILGFFKGLQAQILKTVLSSALLLMIKEKITATTWILILAIRTLFVTKAR 317
> YIL006w
Length=373
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 47/158 (29%), Positives = 76/158 (48%), Gaps = 36/158 (22%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
F+ AG+VP+L+ + + AI F +Y+ LK R+ SRE ++NS++ Q S
Sbjct 233 FKALYAGLVPSLLGLFHVAIHFPIYE-------DLKVRFHCYSRENNTNSINLQRLIMAS 285
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLI 119
VS K+ A+ VTYP+ +++TR Q K D S+ R L
Sbjct 286 SVS--------------------KMIASAVTYPHEILRTRMQLK----SDIPDSIQRRLF 321
Query 120 MILET----EGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+++ EGL G ++G + L T+ +SAI +E
Sbjct 322 PLIKATYAQEGLKGFYSGFTTNLVRTIPASAITLVSFE 359
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (41%), Gaps = 13/148 (8%)
Query 18 PAIQFTLYDHLRAKLMQLKQR-WRTASREPSSNSLSS---QHSSTGSLVSDRMLRIRVTP 73
P Q+ DH L+ +Q + S + SS + +SST S++ LR + P
Sbjct 15 PEQQYCSADHEEPLLLHEEQLIFPDHSSQLSSADIIEPIKMNSSTESIIGT-TLRKKWVP 73
Query 74 AESF----LMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILET----E 125
S L G A + + P V KTR Q++ + + R ++ L T E
Sbjct 74 LSSTQITALSGAFAGFLSGVAVCPLDVAKTRLQAQGLQTRFENPYYRGIMGTLSTIVRDE 133
Query 126 GLSGLFAGLPSKLTATLLSSAIMFTVYE 153
G GL+ GL + + I F+VYE
Sbjct 134 GPRGLYKGLVPIVLGYFPTWMIYFSVYE 161
> At2g47490_2
Length=312
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 40/153 (26%), Positives = 72/153 (47%), Gaps = 29/153 (18%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG +G+VPAL + + AIQF Y+ ++ L + + S ++L+++ + S
Sbjct 170 RGLYSGLVPALAGISHVAIQFPTYEMIKVYLAKKGDK--------SVDNLNARDVAVASS 221
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLI-M 120
+ AK+ A+ +TYP+ VV+ R Q + ++ + S VR I
Sbjct 222 I--------------------AKIFASTLTYPHEVVRARLQEQGHHSEKRYSGVRDCIKK 261
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+ E +G G + G + L T ++ I FT +E
Sbjct 262 VFEKDGFPGFYRGCATNLLRTTPAAVITFTSFE 294
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 51/159 (32%), Positives = 72/159 (45%), Gaps = 24/159 (15%)
Query 2 RGWLAGIVPALVLVL-NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSL--SSQHSST 58
RG G+ P ++ +L N AI FT+YD L++ L SN L S+Q+
Sbjct 57 RGLYRGLSPTVMALLSNWAIYFTMYDQLKSFL--------------CSNGLCFSTQNLVN 102
Query 59 GSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVK--DYSSSVR 116
L D L + + L A A + T P VVKTR Q++ V Y S+
Sbjct 103 SCLSGDHKLSV----GANVLAASGAGAATTIATNPLWVVKTRLQTQGMRVGIVPYKSTFS 158
Query 117 SLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+L I EG+ GL++GL L A + AI F YE +
Sbjct 159 ALRRIAYEEGIRGLYSGLVPAL-AGISHVAIQFPTYEMI 196
Score = 35.8 bits (81), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 115 VRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
V SL I + EG+ GL+ GL + A L + AI FT+Y+QL
Sbjct 44 VGSLEQIFKREGMRGLYRGLSPTVMALLSNWAIYFTMYDQL 84
> At3g05290
Length=322
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 47/170 (27%), Positives = 82/170 (48%), Gaps = 31/170 (18%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G+ +L+L NPAIQ+T++D L+ L LKQ+ + +++ S+ ++S M
Sbjct 160 GLGISLLLTSNPAIQYTVFDQLKQHL--LKQK-----------NAKAENGSSPVVLSAFM 206
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ----SKVYNVKDYSSSVRSLI--- 119
+F++G +K A ++TYP + K Q SK K R I
Sbjct 207 ---------AFVLGAVSKSVATVLTYPAIRCKVMIQAADESKENETKKPRRRTRKTIPGV 257
Query 120 --MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLR 167
I EG+ G F GL +++ T+LSSA++ + E++ +L ++R
Sbjct 258 VYAIWRKEGMLGFFKGLQAQILKTVLSSALLLMIKEKITATTWILILAIR 307
> YIL134w
Length=311
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 44/156 (28%), Positives = 68/156 (43%), Gaps = 29/156 (18%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
F+G G+VPAL V A+ F +YD LKQR RE
Sbjct 180 FQGLWKGLVPALFGVSQGALYFAVYD-------TLKQRKLRRKRENG------------- 219
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLI- 119
L I +T E+ + K+ + + YP+ ++K+ QS + + + LI
Sbjct 220 ------LDIHLTNLETIEITSLGKMVSVTLVYPFQLLKSNLQS--FRANEQKFRLFPLIK 271
Query 120 MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+I+ +G GL+ GL + L + S+ I F VYE L
Sbjct 272 LIIANDGFVGLYKGLSANLVRAIPSTCITFCVYENL 307
Score = 36.6 bits (83), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKD-YSSSVRSLIMILETEGLSGLFAGLPSKL 138
G ++ L A++T P V+KTR S + Y+S + +L T+G GL+ GL L
Sbjct 132 GASSGLMTAILTNPIWVIKTRIMSTSKGAQGAYTSMYNGVQQLLRTDGFQGLWKGLVPAL 191
Query 139 TATLLSSAIMFTVYEQL 155
+ A+ F VY+ L
Sbjct 192 FG-VSQGALYFAVYDTL 207
> YGR257c
Length=366
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 43/85 (50%), Gaps = 6/85 (7%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIM------ILETEGLS 128
SF G + + AA+ T+P+ V KTR Q + N D RS M I TEGL+
Sbjct 270 NSFASGCISGMIAAICTHPFDVGKTRWQISMMNNSDPKGGNRSRNMFKFLETIWRTEGLA 329
Query 129 GLFAGLPSKLTATLLSSAIMFTVYE 153
L+ GL +++ S AIM + YE
Sbjct 330 ALYTGLAARVIKIRPSCAIMISSYE 354
> YBR192w
Length=377
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 48/79 (60%), Gaps = 2/79 (2%)
Query 83 AKLAAALVTYPYLVVKTRAQS--KVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTA 140
AK A++ TYP+ VV+TR + K + Y+ V+S +I++ EGL +++GL L
Sbjct 298 AKFVASIATYPHEVVRTRLRQTPKENGKRKYTGLVQSFKVIIKEEGLFSMYSGLTPHLMR 357
Query 141 TLLSSAIMFTVYEQLLPLV 159
T+ +S IMF +E ++ L+
Sbjct 358 TVPNSIIMFGTWEIVIRLL 376
Score = 32.7 bits (73), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 93 PYLVVKTRAQ---SKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMF 149
P ++KTR Q + +V+ Y +S L ++ EG +GL+ GL + + + + +
Sbjct 195 PIWLIKTRVQLDKAGKTSVRQYKNSWDCLKSVIRNEGFTGLYKGLSASYLGS-VEGILQW 253
Query 150 TVYEQLLPLV 159
+YEQ+ L+
Sbjct 254 LLYEQMKRLI 263
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 45/108 (41%), Gaps = 26/108 (24%)
Query 71 VTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVY---------NVKDYSSSVRSLIMI 121
V P F+ G +A A+VT P+ +VKTR QS ++ N+ S+ +S+ +
Sbjct 50 VKPWVHFVAGGIGGMAGAVVTCPFDLVKTRLQSDIFLKAYKSQAVNISKGSTRPKSINYV 109
Query 122 LET-----------------EGLSGLFAGLPSKLTATLLSSAIMFTVY 152
++ EG LF GL L + + +I F Y
Sbjct 110 IQAGTHFKETLGIIGNVYKQEGFRSLFKGLGPNLVGVIPARSINFFTY 157
> YEL006w
Length=335
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 43/157 (27%), Positives = 78/157 (49%), Gaps = 33/157 (21%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
+ AG+VPAL+ +LN AIQF LY++L+ + + S + S++ SS L
Sbjct 196 KALYAGLVPALLGMLNVAIQFPLYENLKIRF------GYSESTDVSTDVTSSNFQ---KL 246
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLIM 120
+ ML +K+ A+ VTYP+ +++TR Q K D ++V R L+
Sbjct 247 ILASML---------------SKMVASTVTYPHEILRTRMQLK----SDLPNTVQRHLLP 287
Query 121 ILE----TEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+++ EG +G ++G + L T+ ++ + +E
Sbjct 288 LIKITYRQEGFAGFYSGFATNLVRTVPAAVVTLVSFE 324
Score = 34.7 bits (78), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 39/78 (50%), Gaps = 6/78 (7%)
Query 81 LAAKLAAALVTYPYLVVKTRAQSK-----VYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
L+ L+A LV P+ V KTR Q++ + + Y + I + EG +GL+ GL
Sbjct 47 LSGALSAMLVC-PFDVAKTRLQAQGLQNMTHQSQHYKGFFGTFATIFKDEGAAGLYKGLQ 105
Query 136 SKLTATLLSSAIMFTVYE 153
+ + + I F+VY+
Sbjct 106 PTVLGYIPTLMIYFSVYD 123
> At2g33820
Length=344
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 42/82 (51%), Gaps = 2/82 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKD--YSSSVRSLIMILETEGLSGLFA 132
+ ++ G+ A LA V +P+ VK + Q +V+ Y + + IL+TEG+ GL+
Sbjct 16 KEYVAGMMAGLATVAVGHPFDTVKVKLQKHNTDVQGLRYKNGLHCASRILQTEGVKGLYR 75
Query 133 GLPSKLTATLLSSAIMFTVYEQ 154
G S S++MF +Y Q
Sbjct 76 GATSSFMGMAFESSLMFGIYSQ 97
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 12/74 (16%)
Query 94 YLVVKTRAQ-----SKVYNVKDYSSSVRSLIMILETEGL-------SGLFAGLPSKLTAT 141
+ V+ R Q S V N + Y+S + + ++ +G+ +G+F G + L
Sbjct 139 HCFVQCRMQIQGTDSLVPNFRRYNSPLDCAVQTVKNDGVDGDIFQVTGIFRGGSATLLRE 198
Query 142 LLSSAIMFTVYEQL 155
+A+ FTVYE L
Sbjct 199 CTGNAVFFTVYEYL 212
> 7296049
Length=358
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 38/159 (23%), Positives = 75/159 (47%), Gaps = 27/159 (16%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
GI + + + F +Y+ +++KL L+QR + +H+ T S
Sbjct 219 GITASYFGICETMVHFVIYEFIKSKL--LEQR-------------NQRHTDTKG--SRDF 261
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEG 126
L E + G +K A+ + YP+ V +TR + + Y+S ++L + + EG
Sbjct 262 L-------EFMMAGAVSKTIASCIAYPHEVARTRLREEG---NKYNSFWQTLHTVWKEEG 311
Query 127 LSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
+GL+ GL ++L + ++AIM YE ++ ++ F +
Sbjct 312 RAGLYRGLATQLVRQIPNTAIMMATYEAVVYVLTRRFNN 350
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 113 SSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQ 154
S V+ L I++ EG LF GL L S AI F Y Q
Sbjct 103 SIVQCLRHIVQNEGPRALFKGLGPNLVGVAPSRAIYFCTYSQ 144
> SPAC19G12.05
Length=291
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 63/143 (44%), Gaps = 30/143 (20%)
Query 17 NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRMLRIRVTPAES 76
N ++FT Y+ ++ L SR P LS+ +
Sbjct 176 NSGVRFTAYNSIKQSLQ---------SRLPPDEKLST--------------------VTT 206
Query 77 FLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPS 136
FL+G A + T P VK+R QS + K+Y +S+ IL +GL ++G
Sbjct 207 FLVGSVAGIITVYCTQPIDTVKSRMQS-LSASKEYKNSIHCAYKILTQDGLLRFWSGATP 265
Query 137 KLTATLLSSAIMFTVYEQLLPLV 159
+L +LS I+FTVYE+++ ++
Sbjct 266 RLARLILSGGIVFTVYEKVMEIL 288
Score = 28.1 bits (61), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 50/111 (45%), Gaps = 21/111 (18%)
Query 50 SLSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAA-LVTYPYLVVKTRAQSKVYNV 108
SLS +H G L R + L GL A +A + LV P+ +KT + + +
Sbjct 93 SLSDEH---GHLTGPRTV----------LAGLGAGVAESVLVLTPFESIKT---AIIDDR 136
Query 109 KDYSSSVRSLI----MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
K + ++ + +I+ G+ GL+ GL + + +S + FT Y +
Sbjct 137 KRPNPRLKGFLQASRIIVHENGIRGLYRGLAATVARQAANSGVRFTAYNSI 187
> At5g48970
Length=339
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 42/161 (26%), Positives = 75/161 (46%), Gaps = 39/161 (24%)
Query 1 FRGWLAGIVPALVLVLN-PAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTG 59
FRG+ G VPAL++V+ +IQFT+ L++ S ++ + H
Sbjct 85 FRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFA--------------SGSTKTEDH---- 126
Query 60 SLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT----RAQSKVYNVKDYSSSV 115
I ++P SF+ G A AA L +YP+ +++T + + KVY ++
Sbjct 127 ---------IHLSPYLSFVSGALAGCAATLGSYPFDLLRTILASQGEPKVY------PTM 171
Query 116 RS-LIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
RS + I+++ G+ GL+ GL L + + + F Y+
Sbjct 172 RSAFVDIIQSRGIRGLYNGLTPTLVEIVPYAGLQFGTYDMF 212
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/163 (25%), Positives = 68/163 (41%), Gaps = 30/163 (18%)
Query 2 RGWLAGIVPALV-LVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
RG G+ P LV +V +QF YD + +M R++ +S+ P +
Sbjct 185 RGLYNGLTPTLVEIVPYAGLQFGTYDMFKRWMMDW-NRYKLSSKIPIN------------ 231
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKV------YNVK----D 110
+ ++ + F+ GL A +A LV +P VVK R Q + Y +
Sbjct 232 ------VDTNLSSFQLFICGLGAGTSAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRA 285
Query 111 YSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
Y + + L I+ +EG GL+ G+ + A+ F YE
Sbjct 286 YRNMLDGLRQIMISEGWHGLYKGIVPSTVKAAPAGAVTFVAYE 328
> YBR291c
Length=299
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
+FL+G + + T P VKTR QS + YSS++ I + EGL + G
Sbjct 217 TFLVGAFSGIVTVYSTMPLDTVKTRMQS--LDSTKYSSTMNCFATIFKEEGLKTFWKGAT 274
Query 136 SKLTATLLSSAIMFTVYEQLL 156
+L +LS I+FT+YE++L
Sbjct 275 PRLGRLVLSGGIVFTIYEKVL 295
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 8/98 (8%)
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+S + + V P SFL G A A A +TYP+ KTR Q + S + R+ +++
Sbjct 1 MSSKATKSDVDPLHSFLAGSLAGAAEACITYPFEFAKTRLQL----IDKASKASRNPLVL 56
Query 122 L----ETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+ +T+G+ ++ G P+ + + I F ++ +
Sbjct 57 IYKTAKTQGIGSIYVGCPAFIIGNTAKAGIRFLGFDTI 94
> At5g64970
Length=428
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 41/151 (27%), Positives = 72/151 (47%), Gaps = 20/151 (13%)
Query 7 GIVPALV-LVLNPAIQFTLYDHLRAKLM---QLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G+VP+LV + + A+ + +YD L++ + + K+R +E + Q
Sbjct 282 GLVPSLVSMAPSGAVFYGVYDILKSAYLHTPEGKKRLEHMKQEGEELNAFDQ-------- 333
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMIL 122
+ + P + L G A + TYP+ VV+ R Q + + + S+V + + I+
Sbjct 334 ------LELGPMRTLLYGAIAGACSEAATYPFEVVRRRLQMQSHAKR--LSAVATCVKII 385
Query 123 ETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
E G+ L+AGL L L S+AI + VYE
Sbjct 386 EQGGVPALYAGLIPSLLQVLPSAAISYFVYE 416
> 7298936
Length=449
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 43/88 (48%), Gaps = 10/88 (11%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYN--------VKDYS--SSVRSLIMILETE 125
SFL G+ A AA+VT P+ VVKT Q + +D+ S+ L I T
Sbjct 328 SFLAGVMAGTVAAIVTTPFDVVKTHEQIEFGERVIFTDSPARDFGKKSTFSRLTGIYRTH 387
Query 126 GLSGLFAGLPSKLTATLLSSAIMFTVYE 153
G+ GLFAG +L + AIM + +E
Sbjct 388 GVRGLFAGCGPRLLKVAPACAIMISTFE 415
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/146 (26%), Positives = 59/146 (40%), Gaps = 32/146 (21%)
Query 40 RTASREPSSNSLSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT 99
R+A + + N L+ SS L+SD +IR P + + + A P V+KT
Sbjct 59 RSAREDDAINRLTDSKSSHRKLLSDPRFQIR--PLQQVISACTGAMITACFMTPLDVIKT 116
Query 100 RAQSK------------------------------VYNVKDYSSSVRSLIMILETEGLSG 129
R QS+ + +SSS +L+ I EGL+
Sbjct 117 RMQSQQSPAHKCFFYSNGLMDHLFASGPNGSELASLRQRPQFSSSWDALMKISRHEGLAA 176
Query 130 LFAGLPSKLTATLLSSAIMFTVYEQL 155
L++GL L + L S+ I F YEQ
Sbjct 177 LWSGLGPTLVSALPSTIIYFVAYEQF 202
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 37/151 (24%), Positives = 68/151 (45%), Gaps = 17/151 (11%)
Query 6 AGIVPALVLVL-NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSD 64
+G+ P LV L + I F Y+ +A+ +Q+ + S+EP + S S+V
Sbjct 179 SGLGPTLVSALPSTIIYFVAYEQFKARYLQIYESHYNKSQEPRHLEIRDTKKSLPSVVP- 237
Query 65 RMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILET 124
+ G+ A++ A V P +V+T+ Q++ VRS++ +
Sbjct 238 ------------MMSGVTARICAVTVVSPIELVRTKMQAQRQTYAQMLQFVRSVVAL--- 282
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+G+ GL+ GL + + S I + +YE L
Sbjct 283 QGVWGLWRGLRPTILRDVPFSGIYWPIYESL 313
> YPR021c
Length=902
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
+F +G A A V YP +KTR Q++ ++ Y +S+ L+ I+ EG+ GL++GL
Sbjct 533 NFSLGSIAGCIGATVVYPIDFIKTRMQAQ-RSLAQYKNSIDCLLKIISREGIKGLYSGLG 591
Query 136 SKLTATLLSSAIMFTV 151
+L AI TV
Sbjct 592 PQLIGVAPEKAIKLTV 607
Score = 31.6 bits (70), Expect = 0.86, Method: Composition-based stats.
Identities = 35/156 (22%), Positives = 63/156 (40%), Gaps = 34/156 (21%)
Query 2 RGWLAGIVPALVLVL-NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
+G +G+ P L+ V AI+ T+ D +R +L
Sbjct 584 KGLYSGLGPQLIGVAPEKAIKLTVNDFMRNRL---------------------------- 615
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKV-YNVKDYSSSVRSLI 119
+D+ ++ + P + G +A + T P +VK R Q + Y ++ + +
Sbjct 616 --TDKNGKLSLFP--EIISGASAGACQVIFTNPLEIVKIRLQVQSDYVGENIQQANETAT 671
Query 120 MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
I++ GL GL+ G+ + L + SAI F Y L
Sbjct 672 QIVKKLGLRGLYNGVAACLMRDVPFSAIYFPTYAHL 707
> YFR045w
Length=178
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 47/92 (51%), Gaps = 3/92 (3%)
Query 65 RMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVK-DYSSSVRSLIMILE 123
R+L+ R A S + GLA +T P VVKTR S+ N K +Y +++ + I
Sbjct 79 RLLQARNDKASSVITGLATSFTLVAMTQPIDVVKTRMMSQ--NAKTEYKNTLNCMYRIFV 136
Query 124 TEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG++ + G + +S + FTVYEQ+
Sbjct 137 QEGMATFWKGSIFRFMKVGISGGLTFTVYEQV 168
> CE01489
Length=328
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 46/80 (57%), Gaps = 2/80 (2%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYN-VKDYSSSVRSLIM-ILETEGLSGLFAG 133
SF+ G AA + A++ T+P+ V+KT Q ++ + D + S+ ++I + + G+S +G
Sbjct 239 SFVSGAAAGVVASIFTHPFDVIKTNCQIRIGGSIDDMNKSITTVIKDMYHSRGISAFSSG 298
Query 134 LPSKLTATLLSSAIMFTVYE 153
L +L S AIM + YE
Sbjct 299 LVPRLVKVSPSCAIMISFYE 318
Score = 28.1 bits (61), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 36/153 (23%), Positives = 61/153 (39%), Gaps = 33/153 (21%)
Query 6 AGIVPALVLVLNPA--IQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVS 63
+G+ P +V+ L PA FT YD+L L + R S E
Sbjct 95 SGLSPTMVMAL-PATVFYFTTYDNLSVWLKKKMCCRRAFSPE------------------ 135
Query 64 DRMLRIRVTPAE---SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIM 120
+ TP + + + G+ A+ A V P +++T+ QSK + VRS
Sbjct 136 ------KWTPPDWSAAAVAGIVARTIAVTVVSPIEMIRTKMQSKRLTYHEIGHLVRSS-- 187
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+ T+G+S + G + + S I + Y+
Sbjct 188 -MATKGISSFYLGWTPTMLRDIPFSGIYWAGYD 219
> At4g39460
Length=330
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 40/77 (51%), Gaps = 5/77 (6%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLT 139
G LAA+L+ P VVK R Q+ ++S+ ++ MI EG GL+AG S L
Sbjct 142 GAIGGLAASLIRVPTEVVKQRMQTG-----QFTSAPSAVRMIASKEGFRGLYAGYRSFLL 196
Query 140 ATLLSSAIMFTVYEQLL 156
L AI F +YEQL
Sbjct 197 RDLPFDAIQFCIYEQLC 213
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 33/79 (41%), Gaps = 15/79 (18%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
E F+ G A + YP +KTR Q+ R I+ L GL++GL
Sbjct 56 EGFIAGGTAGVVVETALYPIDTIKTRLQA-----------ARGGGKIV----LKGLYSGL 100
Query 135 PSKLTATLLSSAIMFTVYE 153
+ L +SA+ VYE
Sbjct 101 AGNIAGVLPASALFVGVYE 119
> YGR096w
Length=314
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 49/82 (59%), Gaps = 4/82 (4%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLIMILETEGLSGLFAG 133
S ++G A + +++V+YP+ V++TR V N + +S S+ R + I + EGL G F G
Sbjct 114 HSLVVGAFAGITSSIVSYPFDVLRTRL---VANNQMHSMSITREVRDIWKLEGLPGFFKG 170
Query 134 LPSKLTATLLSSAIMFTVYEQL 155
+ +T L+++IMF YE +
Sbjct 171 SIASMTTITLTASIMFGTYETI 192
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 13/87 (14%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQ-------------SKVYNVKDYSSSVRSLIMILETEG 126
G + A ++T+P ++ R Q S VY R + IL+ EG
Sbjct 219 GTIGGVIAKIITFPLETIRRRMQFMNSKHLEKFSRHSSVYGSYKGYGFARIGLQILKQEG 278
Query 127 LSGLFAGLPSKLTATLLSSAIMFTVYE 153
+S L+ G+ L+ T+ ++ + F YE
Sbjct 279 VSSLYRGILVALSKTIPTTFVSFWGYE 305
> Hs21314739
Length=315
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 39/154 (25%), Positives = 60/154 (38%), Gaps = 30/154 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG G VP L + A+QF Y+ L+ K Q +R P + + ++ S +L
Sbjct 180 RGLYKGFVPGLFGTSHGALQFMAYELLKLKYNQ------HINRLPEAQLSTVEYISVAAL 233
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+K+ A TYPY VV+ R Q + YS + +
Sbjct 234 ---------------------SKIFAVAATYPYQVVRARLQDQ---HMFYSGVIDVITKT 269
Query 122 LETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ G + G+ L + I F VYE +
Sbjct 270 WRKEGVGGFYKGIAPNLIRVTPACCITFVVYENV 303
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTR-AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFA 132
E+ + G++ + + L +P +VK R A S ++ Y+ + L I + +GL GL+
Sbjct 24 ENLIAGVSGGVLSNLALHPLDLVKIRFAVSDGLELRPKYNGILHCLTTIWKLDGLRGLYQ 83
Query 133 GLPSKLTATLLSSAIMFTVYEQL 155
G+ + LS + F Y +
Sbjct 84 GVTPNIWGAGLSWGLYFFFYNAI 106
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 36/91 (39%), Gaps = 6/91 (6%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNV-----KDYSSSVRSLIMILET 124
R+ E + A +T P V KTR + V + Y +L+ I +
Sbjct 117 RLEATEYLVSAAEAGAMTLCITNPLWVTKTRLMLQYDAVVNSPHRQYKGMFDTLVKIYKY 176
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ GL+ G L T A+ F YE L
Sbjct 177 EGVRGLYKGFVPGLFGT-SHGALQFMAYELL 206
> HsM13540551
Length=315
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 39/154 (25%), Positives = 60/154 (38%), Gaps = 30/154 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG G VP L + A+QF Y+ L+ K Q +R P + + ++ S +L
Sbjct 180 RGLYKGFVPGLFGTSHGALQFMAYELLKLKYNQ------HINRLPEAQLSTVEYISVAAL 233
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+K+ A TYPY VV+ R Q + YS + +
Sbjct 234 ---------------------SKIFAVAATYPYQVVRARLQDQ---HMFYSGVIDVITKT 269
Query 122 LETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ G + G+ L + I F VYE +
Sbjct 270 WRKEGVGGFYKGIAPNLIRVTPACCITFVVYENV 303
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTR-AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFA 132
E+ + G++ + + L +P +VK R A S ++ Y+ + L I + +GL GL+
Sbjct 24 ENLIAGVSGGVLSNLALHPLDLVKIRFAVSDGLELRPKYNGILHCLTTIWKLDGLRGLYQ 83
Query 133 GLPSKLTATLLSSAIMFTVYEQL 155
G+ + LS + F Y +
Sbjct 84 GVTPNIWGAGLSWGLYFFFYNAI 106
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 38/88 (43%), Gaps = 7/88 (7%)
Query 74 AESFLMGLAAKLAAAL-VTYPYLVVKTRAQSKVYNV-----KDYSSSVRSLIMILETEGL 127
A +L+ A A L +T P V KTR + V + Y +L+ I + EG+
Sbjct 120 ATEYLVSAAEAGAMTLCITNPLWVTKTRLMLQYDAVVNSPHRQYKGMFDTLVKIYKYEGV 179
Query 128 SGLFAGLPSKLTATLLSSAIMFTVYEQL 155
GL+ G L T A+ F YE L
Sbjct 180 RGLYKGFVPGLFGT-SHGALQFMAYELL 206
> Hs8923520
Length=304
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 51/97 (52%), Gaps = 10/97 (10%)
Query 59 GSLVSDRMLRIRVTPA---ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV 115
G+L S + +R P ES ++G+ ++ A + P V+KTR +S Y + +++
Sbjct 106 GTLYSLKQYFLRGHPPTALESVMLGVGSRSVAGVCMSPITVIKTRYESGKYGYESIYAAL 165
Query 116 RSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVY 152
RS I +EG GLF+G LTATLL A +Y
Sbjct 166 RS---IYHSEGHRGLFSG----LTATLLRDAPFSGIY 195
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 59/103 (57%), Gaps = 5/103 (4%)
Query 55 HSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSS 114
++ T ++V + + P +F G+ A + A+LVT P V+KT Q +Y +K +
Sbjct 199 YNQTKNIVPHDQVDATLIPITNFSCGIFAGILASLVTQPAGVIKTHMQ--LYPLK-FQWI 255
Query 115 VRSLIMILETEGLSGLF-AGLPSKLTATLLSSAIMFTVYEQLL 156
+++ +I + GL G F G+P L TL+ +A+ +TVYE+++
Sbjct 256 GQAVTLIFKDYGLRGFFQGGIPRALRRTLM-AAMAWTVYEEMM 297
> At3g55640
Length=332
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 34/81 (41%), Positives = 44/81 (54%), Gaps = 5/81 (6%)
Query 77 FLMGLAAKLAAALVTYPYLVVKTR--AQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
F+ G A + AA TYP +V+TR AQ+KV YS +L I EG+ GL+ GL
Sbjct 145 FVAGGLAGITAASATYPLDLVRTRLAAQTKVIY---YSGIWHTLRSITTDEGILGLYKGL 201
Query 135 PSKLTATLLSSAIMFTVYEQL 155
+ L S AI F+VYE L
Sbjct 202 GTTLVGVGPSIAISFSVYESL 222
> Hs18583647
Length=303
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 39/83 (46%), Gaps = 0/83 (0%)
Query 74 AESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAG 133
A FL G A +A LV +P+ VK R Q + Y ++ I++ E + GL+ G
Sbjct 2 ALDFLAGCAGGVAGVLVGHPFDTVKVRLQVQSVEKPQYRGTLHCFKSIIKQESVLGLYKG 61
Query 134 LPSKLTATLLSSAIMFTVYEQLL 156
L S L +A++F V L
Sbjct 62 LGSPLMGLTFINALVFGVQGNTL 84
> SPAC688.09
Length=361
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/161 (22%), Positives = 72/161 (44%), Gaps = 20/161 (12%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
FRG G+ +L+ V +Q+ LY+ + + A R+ L Q +
Sbjct 218 FRGLYKGLSASLLGVGESTLQWVLYEKFKHAV---------AIRQLRRKELGIQET---- 264
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSK--VYNVKDYSSSVRSL 118
+ D++L + + AK AA + YP+ VV+TR + + Y+ ++
Sbjct 265 -IYDKVLDWGGKLGGAGI----AKFMAAGIAYPHEVVRTRLRQSPSINGTPKYTGLIQCF 319
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLV 159
++ +G+ GL+ GL + L + ++ I+F YE ++ +
Sbjct 320 KLVWMEQGIVGLYGGLTAHLLRVVPNACILFGSYEVIMHFI 360
Score = 36.2 bits (82), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 44/92 (47%), Gaps = 8/92 (8%)
Query 81 LAAKLAAAL---VTYPYLVVKTRAQ--SKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
+AA +A + T P +VKTR Q K Y SS+ +I + EG GL+ GL
Sbjct 167 MAAAIAGVITSAATNPIWLVKTRLQLDKKSGQAAQYRSSIDCIIKTIRLEGFRGLYKGLS 226
Query 136 SKLTATLLSSAIMFTVYEQLLPLVVLLFRSLR 167
+ L + S + + +YE+ V + R LR
Sbjct 227 ASLLG-VGESTLQWVLYEKFKHAVAI--RQLR 255
> SPBC27B12.09c
Length=277
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 66/158 (41%), Gaps = 46/158 (29%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG AG P+L+ V A+QF Y+ KL KQR
Sbjct 150 RGCYAGFAPSLLGVSQGALQFMAYE----KLKLWKQR----------------------- 182
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLI-M 120
R T + M A+K+ AA+ YP LV++TR Q S RS++ +
Sbjct 183 --------RPTSLDYIFMSAASKVFAAVNMYPLLVIRTRLQVM-------RSPHRSIMNL 227
Query 121 ILET---EGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+L+T +G+ G + G L + + I F VYEQ+
Sbjct 228 VLQTWRLQGILGFYKGFLPHLLRVVPQTCITFLVYEQV 265
> SPBC29A3.11c
Length=297
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 44/84 (52%), Gaps = 5/84 (5%)
Query 72 TPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLF 131
+PA +F L + + ++ YP +K R Q+ YN S ++ + + EGL GL+
Sbjct 16 SPASTFSAALVSSAISNVIGYPLDSIKVRQQT--YNFPTIRSCFQNAV---KNEGLKGLY 70
Query 132 AGLPSKLTATLLSSAIMFTVYEQL 155
GL L + LS ++ FTVY+ L
Sbjct 71 RGLTLPLISATLSRSVSFTVYDSL 94
> YKL120w
Length=324
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQS-----KVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
G A+ + A++ P +VKTR QS K+ Y+ L+ I +TEG+ GLF G+
Sbjct 135 GAASGIIGAVIGSPLFLVKTRLQSYSEFIKIGEQTHYTGVWNGLVTIFKTEGVKGLFRGI 194
Query 135 PSKLTATLLSSAIMFTVYE 153
+ + T S++ +Y
Sbjct 195 DAAILRTGAGSSVQLPIYN 213
> Hs8922551
Length=310
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/165 (24%), Positives = 72/165 (43%), Gaps = 29/165 (17%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASR-EPSSNSLSSQHSSTGS 60
+G+ G+ + + I F +Y+ ++ KL++ ++TAS E S+ G
Sbjct 174 KGFYRGMSASYAGISETVIHFVIYESIKQKLLE----YKTASTMENGEESVKEASDFVGM 229
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIM 120
+ L +K A + YP+ VV+TR + + Y S ++L +
Sbjct 230 M----------------LAAATSKTCATTIAYPH-VVRTRLREEG---TKYRSFFQTLSL 269
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
+++ EG L+ GL + L + ++AIM YE LVV L
Sbjct 270 LVQEEGYGSLYRGLTTHLVRQIPNTAIMMATYE----LVVYLLNG 310
Score = 33.1 bits (74), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 36/95 (37%), Gaps = 21/95 (22%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVR-------------------SLIM 120
G A++T P VVKTR QS +V Y S V+ L +
Sbjct 13 GGCGGTVGAILTCPLEVVKTRLQSS--SVTLYISEVQLNTMAGASVNRVVSPGPLHCLKV 70
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
ILE EG LF GL L S AI F Y
Sbjct 71 ILEKEGPRSLFRGLGPNLVGVAPSRAIYFAAYSNC 105
> CE10358
Length=294
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 45/88 (51%), Gaps = 3/88 (3%)
Query 78 LMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLS-GLFAGLPS 136
L G+ A L +YP +V+ R Q+ + S +R+LI I TEGL GL+ GL
Sbjct 209 LFGMLAGLIGQSSSYPLDIVRRRMQTG--RIPSGWSPLRALIHIYHTEGLKRGLYKGLSM 266
Query 137 KLTATLLSSAIMFTVYEQLLPLVVLLFR 164
++ + FT YE++L LV L R
Sbjct 267 NWLKGPIAVGVSFTTYEKVLELVGHLKR 294
> CE02342
Length=384
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/155 (23%), Positives = 64/155 (41%), Gaps = 35/155 (22%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
F+G+ G+ + V IQF +Y++ R G
Sbjct 254 FKGFYKGVTASYAGVSETMIQFCIYEYFR-----------------------------GV 284
Query 61 LVSD--RMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSL 118
L+SD M + ++ + G +AK A +V YP+ VV+TR + + + + ++ L
Sbjct 285 LLSDANEMDKRKMDFLNFMVAGGSAKFIACVVAYPHEVVRTRLREETGKSRGFFKTLYQL 344
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
EG ++ GL +L T+ ++AI YE
Sbjct 345 Y----KEGYPAMYRGLSVQLMRTVPNTAITMGTYE 375
> SPCC1682.09c
Length=300
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 53/120 (44%), Gaps = 2/120 (1%)
Query 39 WRTASREPSSNSLSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVK 98
W A P S +L ++ + + T ++F +A A+ +V+ P V+K
Sbjct 177 WTAARNAPGSFALFGGNAFAKEYIFKLKDYSQATFFQNFFTSIAGASASLIVSAPLDVIK 236
Query 99 TRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPL 158
TR Q+K N + S L +L+ EG + F GL KL T FT+ + L+P
Sbjct 237 TRIQNK--NFDNPQSGFTILKNMLKFEGPTSFFKGLTPKLLTTGPKLVFSFTMAQTLIPF 294
> 7303925
Length=360
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 49/204 (24%), Positives = 70/204 (34%), Gaps = 64/204 (31%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG G VP ++ V + AIQF Y+ +LK + + P L++
Sbjct 179 RGLYRGFVPGMLGVSHGAIQFMTYE-------ELKNAYNEYRKLPIDTKLAT-------- 223
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKD----------- 110
E +KL AA TYPY VV+ R Q +
Sbjct 224 ------------TEYLAFAAVSKLIAAAATYPYQVVRARLQDHHHRYNGTWDCIKQTWRY 271
Query 111 ------YSSSVRSLIMI------------------LETEGLSGLFAGLPSKLTATLLSSA 146
Y V L+ + LE EG G + GL + LT + +
Sbjct 272 ERMRGFYKGLVPYLVHVTPNICMPASFHLSKGSWQLEFEGYRGFYKGLKASLTRVVPACM 331
Query 147 IMFTVYEQLLPLVVLLFRSLRVPT 170
+ F VYE + LL R R+ T
Sbjct 332 VTFLVYENVSHF--LLARRKRIET 353
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 5/86 (5%)
Query 73 PAESFLMGLAAKLAAALVTYPYLVVKTRA--QSKVYNVKDYSSSVRSLIMILETEGLSGL 130
P + L + + L+T P VVKTR Q + +Y + +L I + EG+ GL
Sbjct 122 PTMNMLAAAESGILTLLLTNPIWVVKTRLCLQCDAASSAEYRGMIHALGQIYKEEGIRGL 181
Query 131 FAGL-PSKLTATLLSSAIMFTVYEQL 155
+ G P L + AI F YE+L
Sbjct 182 YRGFVPGMLGVS--HGAIQFMTYEEL 205
> Hs10047122
Length=338
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 46/85 (54%), Gaps = 7/85 (8%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-----RSLIM--ILETEGLS 128
+F G + AA+ T P+ VVKT+ Q++++ + + S+ +IM I+ G S
Sbjct 238 NFTSGALSGSFAAVATLPFDVVKTQKQTQLWTYESHKISMPLHMSTWIIMKNIVAKNGFS 297
Query 129 GLFAGLPSKLTATLLSSAIMFTVYE 153
GLF+GL +L + AIM + YE
Sbjct 298 GLFSGLIPRLIKIAPACAIMISTYE 322
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 27/118 (22%), Positives = 49/118 (41%), Gaps = 31/118 (26%)
Query 69 IRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSK----------VYNVK--------- 109
I+VTP + L + +++ P VVK R Q++ VY+
Sbjct 11 IKVTPLQQMLASCTGAILTSVIVTPLDVVKIRLQAQNNPLPKGKCFVYSNGLMDHLCVCE 70
Query 110 ------------DYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
++ ++ + I+ EG+ L++GLP L + ++ I FT Y+QL
Sbjct 71 EGGNKLWYKKPGNFQGTLDAFFKIIRNEGIKSLWSGLPPTLVMAVPATVIYFTCYDQL 128
> At3g21390
Length=327
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 40/78 (51%), Gaps = 7/78 (8%)
Query 77 FLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGL-P 135
FL GLA+ + LV +P VVK R + + Y + L IL +EG GL+ G+ P
Sbjct 245 FLCGLASGTVSKLVCHPLDVVKKR-----FQLNAYKNMFDGLGQILRSEGWHGLYKGIVP 299
Query 136 SKLTATLLSSAIMFTVYE 153
S + A + A+ F YE
Sbjct 300 STIKAA-PAGAVTFVAYE 316
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 49/89 (55%), Gaps = 11/89 (12%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKT----RAQSKVYNVKDYSSSVRSLIM-ILET 124
+++P S++ G A AA + +YP+ +++T + + KVY ++RS + I++T
Sbjct 134 QLSPYLSYISGALAGCAATVGSYPFDLLRTVLASQGEPKVY------PNMRSAFLSIVQT 187
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
G+ GL+AGL L + + + F Y+
Sbjct 188 RGIKGLYAGLSPTLIEIIPYAGLQFGTYD 216
> At1g14560
Length=331
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 45/162 (27%), Positives = 66/162 (40%), Gaps = 41/162 (25%)
Query 2 RGWLAGIVPALVLVLNPA-IQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
RG GI P L+ +L A ++F +Y+ L+ + + Q NS+ H G+
Sbjct 188 RGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQ-----------NSVR-MHLPCGA 235
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ-------SKVYNVKDYSS 113
L A L +TYP VV+ + Q + N K Y +
Sbjct 236 L---------------------AGLFGQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKN 274
Query 114 SVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+ L I+ T+G LFAGL + S AI FTVYE +
Sbjct 275 TFDGLNTIVRTQGWKQLFAGLSINYIKIVPSVAIGFTVYESM 316
Score = 34.7 bits (78), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 41/106 (38%), Gaps = 14/106 (13%)
Query 64 DRMLRIRVTPAESFLMGLAAKLAAALVTYP---------YLVVKTR-----AQSKVYNVK 109
++ L + P + G AA A L TYP Y V TR + Y
Sbjct 110 EKNLPLGSGPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQP 169
Query 110 DYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
YS L M + G GL+ G+ L L + + F +YE+L
Sbjct 170 TYSGIKEVLAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEEL 215
> YDL198c
Length=300
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 55/119 (46%), Gaps = 2/119 (1%)
Query 39 WRTASREPSSNSLSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVK 98
W A P S +L ++ + + T +++F+ + ++ +V+ P V+K
Sbjct 176 WTAARNAPGSFALFGGNAFAKEYILGLKDYSQATWSQNFISSIVGACSSLIVSAPLDVIK 235
Query 99 TRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLP 157
TR Q++ N + S +R + L+ EG++ F GL KL T F + + L+P
Sbjct 236 TRIQNR--NFDNPESGLRIVKNTLKNEGVTAFFKGLTPKLLTTGPKLVFSFALAQSLIP 292
> At4g01100
Length=352
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 106 YNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFR 164
+N+K YS +V+ L I TEGL GLF G + + +SA+ F YEQ ++ ++R
Sbjct 72 HNIK-YSGTVQGLKHIWRTEGLRGLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYR 129
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 36/87 (41%), Gaps = 1/87 (1%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVK-DYSSSVRSLIMILETEGLS 128
++TP G A + A TYP +V+ R + N Y +L +L EG
Sbjct 138 QLTPLLRLGAGATAGIIAMSATYPMDMVRGRLTVQTANSPYQYRGIAHALATVLREEGPR 197
Query 129 GLFAGLPSKLTATLLSSAIMFTVYEQL 155
L+ G + + + F+VYE L
Sbjct 198 ALYRGWLPSVIGVVPYVGLNFSVYESL 224
> At2g37890
Length=348
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 39/84 (46%), Gaps = 1/84 (1%)
Query 72 TPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLF 131
P F+ G A + AA TYP +V+TR ++ N Y + I EG+ GL+
Sbjct 147 NPIVHFVSGGLAGITAATATYPLDLVRTRLAAQ-RNAIYYQGIEHTFRTICREEGILGLY 205
Query 132 AGLPSKLTATLLSSAIMFTVYEQL 155
GL + L S AI F YE +
Sbjct 206 KGLGATLLGVGPSLAINFAAYESM 229
> Hs21389315
Length=311
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 46/95 (48%), Gaps = 2/95 (2%)
Query 71 VTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGL 130
+ P + + G A A+ P V+KTR Q Y ++ + IL+ EGL
Sbjct 218 MNPLITGVFGAIAGAASVFGNTPLDVIKTRMQG--LEAHKYRNTWDCGLQILKKEGLKAF 275
Query 131 FAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
+ G +L L AI+F +Y++++ L+ ++++
Sbjct 276 YKGTVPRLGRVCLDVAIVFVIYDEVVKLLNKVWKT 310
> At5g58970
Length=295
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 52/119 (43%), Gaps = 7/119 (5%)
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ-------SKVYNVKDYSSS 114
++D RI ++ E+F+ A A L T P K R Q N+ Y S
Sbjct 1 MADFKPRIEISFLETFICSAFAACFAELCTIPLDTAKVRLQLQRKIPTGDGENLPKYRGS 60
Query 115 VRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLRVPTYRK 173
+ +L I EG+SGL+ G+ + L + + +YE + L+V +P Y+K
Sbjct 61 IGTLATIAREEGISGLWKGVIAGLHRQCIYGGLRIGLYEPVKTLLVGSDFIGDIPLYQK 119
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 35/78 (44%), Gaps = 4/78 (5%)
Query 78 LMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSK 137
L GLAA A + P VVK+R Y ++V I ++TEG+ + G
Sbjct 221 LAGLAAGFFAVCIGSPIDVVKSRMMGD----STYRNTVDCFIKTMKTEGIMAFYKGFLPN 276
Query 138 LTATLLSSAIMFTVYEQL 155
T +AIMF EQ+
Sbjct 277 FTRLGTWNAIMFLTLEQV 294
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 47/99 (47%), Gaps = 13/99 (13%)
Query 81 LAAKLAAAL---VTYPYLVVKTRAQSK----VYNVKDYSSSVRSLIMILETEGLSGLFAG 133
LAA L A+ V P +VK R QS+ + Y+ +V + I++ EG+S L+ G
Sbjct 121 LAALLTGAIAIIVANPTDLVKVRLQSEGKLPAGVPRRYAGAVDAYFTIVKLEGVSALWTG 180
Query 134 LPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLRVPTYR 172
L + + +A Y+Q+ + +++P +R
Sbjct 181 LGPNIARNAIVNAAELASYDQIKETI------MKIPFFR 213
> 7297045
Length=337
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 51/114 (44%), Gaps = 10/114 (8%)
Query 48 SNSLSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ----- 102
NS + LV+++ + P E +L A+ +A +V YP+ + KTR Q
Sbjct 15 DNSEEKERPKLEYLVTNK----KTPPVELYLTAFASACSAEIVGYPFDMCKTRMQIQGEI 70
Query 103 -SKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
S+V Y + + + I+ EGL L+ G+ + L L S I Y+ +
Sbjct 71 ASRVGQKAKYRGLLATAMGIVREEGLLKLYGGISAMLFRHSLFSGIKMLTYDYM 124
> 7297659
Length=399
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 0/80 (0%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
F+ GL A LV +P+ VK Q+ Y + I++ + GL+ G+
Sbjct 43 DFVAGLLGGAAGVLVGHPFDTVKVHLQTDDPRNPKYKGTFHCFRTIVQRDKFIGLYRGIS 102
Query 136 SKLTATLLSSAIMFTVYEQL 155
S + L +AI+F VY +
Sbjct 103 SPMGGIGLVNAIVFGVYGNV 122
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query 77 FLMGLAAKLAAALVTYPYLVVKTRAQ--SKVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
F G A +A V P + KTR Q ++V + ++ + L I++TEG+ G F GL
Sbjct 136 FFAGSIAGVAQGFVCAPMELAKTRLQLSTQVDSGIKFTGPIHCLKYIVKTEGIRGAFKGL 195
Query 135 PSKLTATLLSSAIMFTVYEQLL 156
+ + + A F +E L+
Sbjct 196 TATILRDIPGFASYFVSFEYLM 217
Score = 32.0 bits (71), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 40/97 (41%), Gaps = 3/97 (3%)
Query 66 MLRIRVTP--AESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVK-DYSSSVRSLIMIL 122
++R TP A + + G A +++ L YP VVKT Q+ Y+ + +
Sbjct 216 LMRQVETPGVAYTLMAGGCAGMSSWLACYPIDVVKTHMQADALGANAKYNGFIDCAMKGF 275
Query 123 ETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLV 159
EG F GL S L +A F V +L +
Sbjct 276 RNEGPQYFFRGLNSTLIRAFPMNAACFFVVSWVLDIC 312
> At5g01340
Length=309
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 49/111 (44%), Gaps = 12/111 (10%)
Query 51 LSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKD 110
L ++H G ++ P +S + G A A T P+ VVKTR ++ + +
Sbjct 197 LWNKHEGDGKILQ---------PWQSMISGFLAGTAGPFCTGPFDVVKTRLMAQSRDSEG 247
Query 111 ---YSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPL 158
Y V ++ I EGL L+ GL +L AIM+ V +Q+ L
Sbjct 248 GIRYKGMVHAIRTIYAEEGLVALWRGLLPRLMRIPPGQAIMWAVADQVTGL 298
Lambda K H
0.322 0.133 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2842629034
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40