bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0305_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
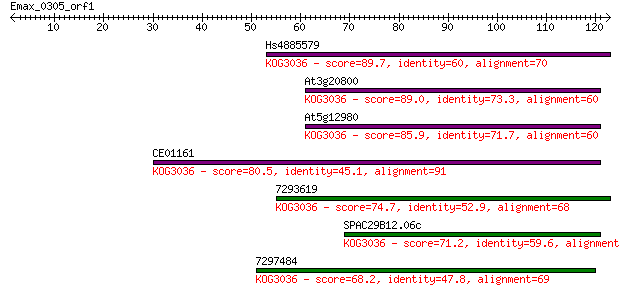
Hs4885579 89.7 1e-18
At3g20800 89.0 2e-18
At5g12980 85.9 2e-17
CE01161 80.5 7e-16
7293619 74.7 4e-14
SPAC29B12.06c 71.2 4e-13
7297484 68.2 4e-12
> Hs4885579
Length=299
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/70 (60%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 53 QQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYP 112
Q +++I Q + +LS E RE ALLELSK+RES PDLAP+LW SFGT+AALLQEI+ YP
Sbjct 16 QVDREKIYQWINELSSPETRENALLELSKKRESVPDLAPMLWHSFGTIAALLQEIVNIYP 75
Query 113 FLSPPSLTTQ 122
++PP+LT
Sbjct 76 SINPPTLTAH 85
> At3g20800
Length=316
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/60 (73%), Positives = 50/60 (83%), Gaps = 0/60 (0%)
Query 61 QLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
QLVLDLS E RE ALLELSK+RE F DLAPLLW SFGT+AALLQEI++ Y L+PP+LT
Sbjct 40 QLVLDLSNPELRENALLELSKKRELFQDLAPLLWNSFGTIAALLQEIVSIYSVLAPPNLT 99
> At5g12980
Length=311
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/60 (71%), Positives = 48/60 (80%), Gaps = 0/60 (0%)
Query 61 QLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
QL+LDLS E RE AL ELSK+RE F DLAPLLW S GT+ ALLQEIIA YP LSPP++T
Sbjct 36 QLILDLSNPELRENALHELSKKREIFQDLAPLLWHSVGTIPALLQEIIAVYPALSPPTMT 95
> CE01161
Length=453
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 41/91 (45%), Positives = 51/91 (56%), Gaps = 0/91 (0%)
Query 30 QQQQQQQQQQQQQQQQQQQQQQQQQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDL 89
+ Q QQ D I Q ++DL KRE ALLELSK+R+S PDL
Sbjct 1 MNDARASPATQAQQTAVPSSANLDINTDEIMQWIIDLRDPPKREAALLELSKKRDSVPDL 60
Query 90 APLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
LW SFGTM+ALLQE++A YP + P +LT
Sbjct 61 PIWLWHSFGTMSALLQEVVAIYPAIMPANLT 91
> 7293619
Length=304
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 51/68 (75%), Gaps = 2/68 (2%)
Query 55 QQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFL 114
+Q+++ Q + +L+ + RE ALLELSK+RE+ DLAP+LW SFGT ALLQEI+ YP +
Sbjct 25 EQEKVYQWINELAHPDTRETALLELSKKRET--DLAPMLWNSFGTACALLQEIVNIYPSI 82
Query 115 SPPSLTTQ 122
+PP+LT
Sbjct 83 TPPTLTAH 90
> SPAC29B12.06c
Length=283
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 31/52 (59%), Positives = 40/52 (76%), Gaps = 0/52 (0%)
Query 69 AEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
RE AL+ELS++RE + DLA +LW S+G M ALLQEII+ YP L+PP+LT
Sbjct 24 GTSREQALVELSRKREQYEDLALILWHSYGVMTALLQEIISVYPLLNPPTLT 75
> 7297484
Length=290
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 48/69 (69%), Gaps = 2/69 (2%)
Query 51 QQQQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAA 110
QQQ +++++ QL+++L+ RE ALLELSK ++ DLAP+LW S GT LLQEI+
Sbjct 12 QQQAEREKVYQLIIELAYPATRETALLELSK--NTYADLAPMLWKSVGTTCTLLQEIVNI 69
Query 111 YPFLSPPSL 119
YP ++ P L
Sbjct 70 YPIITTPVL 78
Lambda K H
0.312 0.121 0.323
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40