bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
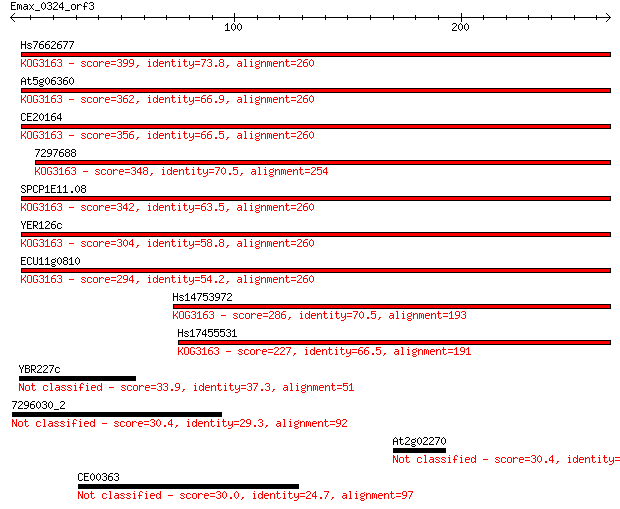
Query= Emax_0324_orf3
Length=265
Score E
Sequences producing significant alignments: (Bits) Value
Hs7662677 399 4e-111
At5g06360 362 6e-100
CE20164 356 3e-98
7297688 348 6e-96
SPCP1E11.08 342 3e-94
YER126c 304 1e-82
ECU11g0810 294 1e-79
Hs14753972 286 3e-77
Hs17455531 227 2e-59
YBR227c 33.9 0.38
7296030_2 30.4 3.6
At2g02270 30.4 4.0
CE00363 30.0 4.7
> Hs7662677
Length=260
Score = 399 bits (1024), Expect = 4e-111, Method: Compositional matrix adjust.
Identities = 192/260 (73%), Positives = 222/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E HRKR+G RLD E+KRKK +R HERSK A+K+ G+KAKL++++R EK Q
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKESREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIK HE++ KQK E P+GAVP+YLLDREG SR K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTIKMHEKRNTKQKNDEKTPQGAVPAYLLDREGQSRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP LRFKK
Sbjct 121 LPKVRAQGETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKV+W K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSPLYTTLGVITKGTVIEVNVSELGLVTQGGKVIWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> At5g06360
Length=260
Score = 362 bits (928), Expect = 6e-100, Method: Compositional matrix adjust.
Identities = 174/260 (66%), Positives = 207/260 (79%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQ +Y++ HRKR G RLD ERKRKK AR H+ S A+K GIK K+ ++ + EKA
Sbjct 1 MPQGDYIDLHRKRNGYRLDHFERKRKKEAREVHKHSTMAQKSLGIKGKMIAKKNYAEKAL 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKT+K HEE ++K E V EGAVP+YLLDRE +R K+L+N IKQKRKEKAGKW+VP
Sbjct 61 MKKTLKMHEESSSRRKADENVQEGAVPAYLLDREDTTRAKVLSNTIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+ + E+EMF+V+RSGKRK K WKR+V K FVG FTRKPPK+ER+IRP+ LRF K
Sbjct 121 LPKVRPVAEDEMFRVIRSGKRKTKQWKRMVTKATFVGPAFTRKPPKYERFIRPSGLRFTK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELK TF L+I+ +KKNP +YTSLGV+T+GTIIEVNVSELGLVT GKVVW K
Sbjct 181 AHVTHPELKCTFCLEIIGIKKNPNGPMYTSLGVMTRGTIIEVNVSELGLVTPAGKVVWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> CE20164
Length=259
Score = 356 bits (914), Expect = 3e-98, Method: Compositional matrix adjust.
Identities = 173/261 (66%), Positives = 214/261 (81%), Gaps = 3/261 (1%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNE++E HRKR G RLD ER+RKK AR H+RS+ A+ LRG KAKL++++R+ EK +
Sbjct 1 MPQNEHIELHRKRHGRRLDHEERQRKKLARAAHDRSQMAKTLRGHKAKLYHKKRYSEKVE 60
Query 66 MKKTIKQHEEKEGKQKEPETVPE-GAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQV 124
M+K +KQHEEK+ QK P+ GAVP+YLLDR+ + +L+NMIKQKRK+KAGK+ V
Sbjct 61 MRKLLKQHEEKD--QKNTVEQPDKGAVPAYLLDRQQQTSGTVLSNMIKQKRKQKAGKFNV 118
Query 125 PIPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFK 184
PIP+V+A+++ E FKV+++GK +K WKR+V KV FVG+ FTRKP KFER+IRP LRFK
Sbjct 119 PIPQVRAVSDAEAFKVVKTGKTNRKGWKRMVTKVTFVGESFTRKPAKFERFIRPMGLRFK 178
Query 185 KAHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 244
KAHVTHPEL+ TF L IV VKKNP SQ+YTSLGVITKGTI+EVNVSELG+VTQ GKVVW
Sbjct 179 KAHVTHPELQTTFHLPIVGVKKNPSSQMYTSLGVITKGTILEVNVSELGMVTQGGKVVWG 238
Query 245 KYAQVTNNPELDGCINAVLLV 265
K+AQVTNNPE DGCINAVLL+
Sbjct 239 KFAQVTNNPENDGCINAVLLI 259
> 7297688
Length=253
Score = 348 bits (893), Expect = 6e-96, Method: Compositional matrix adjust.
Identities = 179/254 (70%), Positives = 212/254 (83%), Gaps = 1/254 (0%)
Query 12 MEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQMKKTIK 71
ME+HRK +G RLD ERKRKK AR+P +R++ ARKLRGIKAKLFN+ R EK Q+KK I+
Sbjct 1 MERHRKLYGRRLDYEERKRKKEARLPKDRARKARKLRGIKAKLFNKERRNEKIQIKKKIQ 60
Query 72 QHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVPIPKVKA 131
HEEK+ K++E + +GA+P YLLDR S K+L+NMIKQKRKEKAGKW VPIPKV+A
Sbjct 61 AHEEKKVKKQEEKVE-DGALPHYLLDRGIQSSAKVLSNMIKQKRKEKAGKWDVPIPKVRA 119
Query 132 MTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKKAHVTHP 191
++ E+FKVL++GK K+K WKR+V KV FVG++FTRKPPKFER+IRP LR KKAHVTHP
Sbjct 120 QSDAEVFKVLKTGKTKRKAWKRMVTKVTFVGENFTRKPPKFERFIRPMGLRMKKAHVTHP 179
Query 192 ELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAKYAQVTN 251
ELKATF L I+ VKKNP S ++TSLGVITKGT+IEVN+SELGLVTQTGKVVW KYAQVTN
Sbjct 180 ELKATFNLPIIGVKKNPSSPMFTSLGVITKGTVIEVNISELGLVTQTGKVVWGKYAQVTN 239
Query 252 NPELDGCINAVLLV 265
NPE DG INAVLLV
Sbjct 240 NPENDGVINAVLLV 253
> SPCP1E11.08
Length=260
Score = 342 bits (878), Expect = 3e-94, Method: Compositional matrix adjust.
Identities = 165/260 (63%), Positives = 203/260 (78%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E+ ++ G R D ERKRKKAAR H+ S A+K RGIKAKL+ +R EK Q
Sbjct 1 MPQNEYIEESIRKHGRRFDHEERKRKKAAREAHDASLYAQKTRGIKAKLYQEKRRKEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIKQHEE+ Q+ + +GAVP+YLLDRE S+ K+L++ +KQKRKEKA K+ VP
Sbjct 61 MKKTIKQHEERNATQRGSDAQTQGAVPTYLLDREQESQAKMLSSAVKQKRKEKAAKYSVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+P+V+ + EEEMFKV+R+GK KK WKR++ K FVGD FTR+P K+ER+IRP +LR KK
Sbjct 121 LPQVRGVAEEEMFKVIRTGKSKKNSWKRMITKATFVGDGFTRRPVKYERFIRPMALRQKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
A+VTH EL T QL I+ VKKNPQS YT LGV+TKGT+IEVNVSELGLVT GKVVW K
Sbjct 181 ANVTHKELGVTMQLPIIGVKKNPQSPTYTQLGVLTKGTVIEVNVSELGLVTSGGKVVWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQ+TNNPELDGC+NA+LL
Sbjct 241 YAQITNNPELDGCVNALLLT 260
> YER126c
Length=261
Score = 304 bits (778), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 153/261 (58%), Positives = 197/261 (75%), Gaps = 1/261 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQN+Y+E+H K+ G+RLD ERKRK+ AR H+ S+ A+KL G K K F ++R+ EK
Sbjct 1 MPQNDYIERHIKQHGKRLDHEERKRKREARESHKISERAQKLTGWKGKQFAKKRYAEKVS 60
Query 66 MKKTIKQHEEKEGK-QKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQV 124
M+K IK HE+ + K +P A+P+YLLDRE + K +++ IKQKR EKA K+ V
Sbjct 61 MRKKIKAHEQSKVKGSSKPLDTDGDALPTYLLDREQNNTAKAISSSIKQKRLEKADKFSV 120
Query 125 PIPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFK 184
P+PKV+ ++EEEMFKV+++GK + K WKR++ K FVG+ FTR+P K ER IRP++LR K
Sbjct 121 PLPKVRGISEEEMFKVIKTGKSRSKSWKRMITKHTFVGEGFTRRPVKMERIIRPSALRQK 180
Query 185 KAHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 244
KA+VTHPEL T L I+ VKKNPQS +YT LGV+TKGTIIEVNVSELG+VT GKVVW
Sbjct 181 KANVTHPELGVTVFLPILAVKKNPQSPMYTQLGVLTKGTIIEVNVSELGMVTAGGKVVWG 240
Query 245 KYAQVTNNPELDGCINAVLLV 265
KYAQVTN P+ DGC+NAVLLV
Sbjct 241 KYAQVTNEPDRDGCVNAVLLV 261
> ECU11g0810
Length=258
Score = 294 bits (752), Expect = 1e-79, Method: Compositional matrix adjust.
Identities = 141/261 (54%), Positives = 195/261 (74%), Gaps = 4/261 (1%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQN+++E+H K++G RLD RK KK R +KTA++L G+KAKLF +++ K Q
Sbjct 1 MPQNDFVERHIKKYGRRLDHELRKYKKERRNEKLVAKTAKRLFGLKAKLFAKKQHAAKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPE-TVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQV 124
+KK +K KE K+K E +V + +P +LLDRE R + + + IKQ+R+EK K+ V
Sbjct 61 LKKDLKM---KESKKKGMEVSVEDNPLPHFLLDREMQERARDINDKIKQQRQEKGSKYSV 117
Query 125 PIPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFK 184
P+PK+ + E+E+F V+ SGKR+ K WKR+V K CFVG DFTRKPPK+ER+IRP ++RFK
Sbjct 118 PLPKIHPLPEKEVFGVVASGKRRSKHWKRMVTKPCFVGSDFTRKPPKYERFIRPMAMRFK 177
Query 185 KAHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 244
AHV+HPEL +TF L I+ +KKNP S++YT LG+++KGT+IEV+V LGLV +GK++W
Sbjct 178 TAHVSHPELNSTFNLKIIGLKKNPHSEVYTGLGILSKGTVIEVDVGPLGLVNGSGKIIWG 237
Query 245 KYAQVTNNPELDGCINAVLLV 265
KYAQ+TNNPE DGC+NAVLLV
Sbjct 238 KYAQITNNPENDGCVNAVLLV 258
> Hs14753972
Length=194
Score = 286 bits (732), Expect = 3e-77, Method: Compositional matrix adjust.
Identities = 136/193 (70%), Positives = 159/193 (82%), Gaps = 0/193 (0%)
Query 73 HEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVPIPKVKAM 132
HE++ KQK E P+GAVP+YLLDRE SR K+L+N IKQKRKEK GKW+VP+PKV+A
Sbjct 2 HEKRNIKQKNDEKTPQGAVPAYLLDREEQSRAKVLSNRIKQKRKEKVGKWEVPLPKVRAQ 61
Query 133 TEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKKAHVTHPE 192
E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP L FKKAHVTHPE
Sbjct 62 GETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLHFKKAHVTHPE 121
Query 193 LKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAKYAQVTNN 252
+KATF L I+ VKK+P S LYT+ VITKGT+IEVN+SELGLV Q GKV+W YAQVTNN
Sbjct 122 MKATFCLRILGVKKSPLSPLYTTFDVITKGTVIEVNMSELGLVMQGGKVIWGNYAQVTNN 181
Query 253 PELDGCINAVLLV 265
PE D CINAVLL+
Sbjct 182 PENDACINAVLLI 194
> Hs17455531
Length=188
Score = 227 bits (579), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 127/191 (66%), Positives = 144/191 (75%), Gaps = 12/191 (6%)
Query 75 EKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVPIPKVKAMTE 134
E K+K E P+GAVP+YLLDREG SR K+L++MIKQKRKEKAGKW+VP+PKV A E
Sbjct 4 ETPKKKKNAEKTPQGAVPAYLLDREGQSRAKVLSSMIKQKRKEKAGKWEVPLPKVCAQGE 63
Query 135 EEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKKAHVTHPELK 194
E+ KV+R+ KRKKK WKRVV VCFV D FTR PPK+ER+IRP LRFKKAHVT+PELK
Sbjct 64 TEVLKVIRTRKRKKKAWKRVVTNVCFVQDGFTRIPPKYERFIRPMGLRFKKAHVTYPELK 123
Query 195 ATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAKYAQVTNNPE 254
TF L I VKKNP S LYT+LGVITKGT +E V+W KYAQVTNNPE
Sbjct 124 VTFCLPIFGVKKNPSSPLYTTLGVITKGTAME------------AAVIWGKYAQVTNNPE 171
Query 255 LDGCINAVLLV 265
DGCINAVLLV
Sbjct 172 NDGCINAVLLV 182
> YBR227c
Length=520
Score = 33.9 bits (76), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 5 KMPQNEYMEQHR---KRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLF 55
K P+N ++Q+ K+FG RL ++ KK A+ + AR LRGI +L
Sbjct 382 KEPKNALLDQYEYIFKQFGVRLCVTQKALKKVAQFALKEGTGARGLRGIMERLL 435
> 7296030_2
Length=1558
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 27/104 (25%), Positives = 48/104 (46%), Gaps = 16/104 (15%)
Query 2 KFAKMPQNEYMEQHRKRFGERLD--------TAERKRKKAARMPHERSKTARKLRGIKAK 53
KF K Q E +Q R + T + KK +R + S+++R+ ++ K
Sbjct 924 KFEKEKQREREQQRRPAMRSSVSSGALAGGSTTQSCLKKVSRF--DTSESSRRATSVRQK 981
Query 54 LFNRR----RFVEKAQMKKTIKQHEEKEGKQKEPETVPEGAVPS 93
NR RF +Q+K T E++ +++EP + G++PS
Sbjct 982 --NRPKISVRFNTVSQIKYTPSAKRERQAREEEPPEMEVGSLPS 1023
> At2g02270
Length=265
Score = 30.4 bits (67), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 10/23 (43%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 170 PKFERYIRPTSLRFKKAHVTHPE 192
P+F R++ PT L++++ +VT PE
Sbjct 173 PRFVRFVGPTDLKYEREYVTRPE 195
> CE00363
Length=1139
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 24/109 (22%), Positives = 44/109 (40%), Gaps = 12/109 (11%)
Query 31 KKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQMKKTIKQHEEKEGK----------- 79
+ R R K R + I + + RR E+ + K +++H +KE
Sbjct 478 SRGNRASSSRGKMTRSVEYIDPDMISERRAAERREQYKLVREHVKKEDGRIEAYGWSLPN 537
Query 80 -QKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVPIP 127
+ E +VP LLD E + T ++ + +++ G+W V P
Sbjct 538 VEAEVSSVPIPVCCRPLLDNEPSLKIWCATGVVLRGGRDERGQWIVGDP 586
Lambda K H
0.317 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5649423594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40