bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
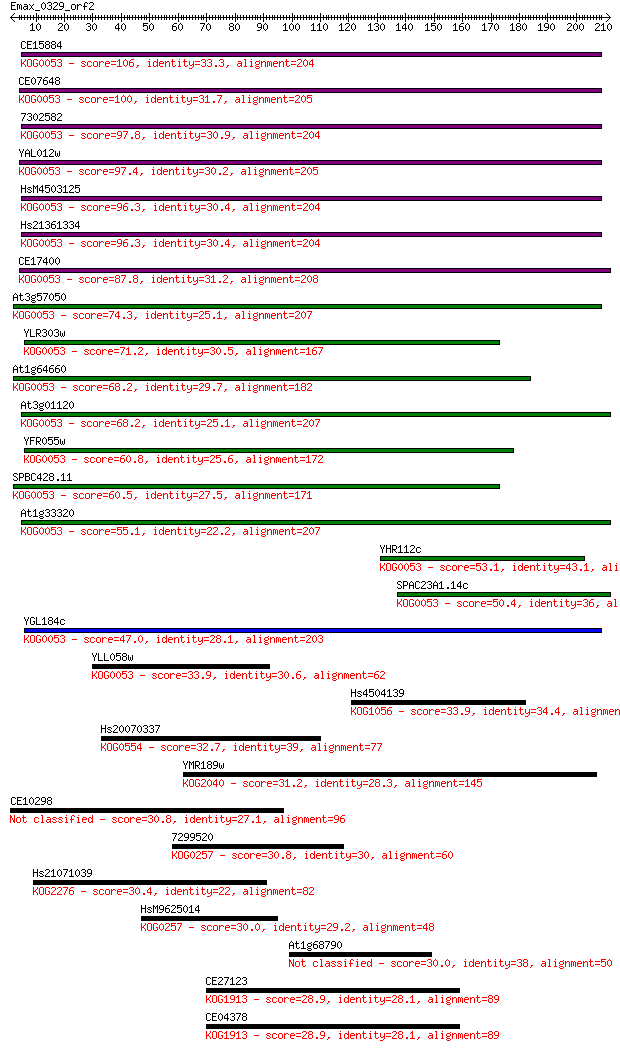
Query= Emax_0329_orf2
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
CE15884 106 3e-23
CE07648 100 3e-21
7302582 97.8 1e-20
YAL012w 97.4 2e-20
HsM4503125 96.3 4e-20
Hs21361334 96.3 4e-20
CE17400 87.8 1e-17
At3g57050 74.3 2e-13
YLR303w 71.2 1e-12
At1g64660 68.2 1e-11
At3g01120 68.2 1e-11
YFR055w 60.8 2e-09
SPBC428.11 60.5 3e-09
At1g33320 55.1 9e-08
YHR112c 53.1 4e-07
SPAC23A1.14c 50.4 3e-06
YGL184c 47.0 3e-05
YLL058w 33.9 0.23
Hs4504139 33.9 0.28
Hs20070337 32.7 0.48
YMR189w 31.2 1.4
CE10298 30.8 1.8
7299520 30.8 2.0
Hs21071039 30.4 3.0
HsM9625014 30.0 3.7
At1g68790 30.0 3.9
CE27123 28.9 7.3
CE04378 28.9 7.3
> CE15884
Length=392
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 68/205 (33%), Positives = 98/205 (47%), Gaps = 41/205 (20%)
Query 5 FSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEY-VSRFGVEFDFVDFRDPSQVE 63
FSSG+AA+ V+ L+ GDH + +YGGT R + V ++G+E D VD D +E
Sbjct 76 FSSGLAASSAVIN-LLNHGDHIVCSDDVYGGTIRYIRQIAVKKYGMEVDSVDLTDVQNLE 134
Query 64 KAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSD 123
KA K NTKMV E+P+NP L + D+ A+ Q + A P
Sbjct 135 KAIKPNTKMVWFESPSNPLLKVVDIAAV--------------VQTAKRANP--------- 171
Query 124 DELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGA 183
I+ VVD+TF TP+ P GADI +HS TKY +GH+ GA
Sbjct 172 ----------------EIVVVVDNTFMTPYFQRPLSLGADIAVHSITKYINGHSDIIMGA 215
Query 184 AISRFLKYHDKIAYTRNICGSIMDP 208
AI+ ++ + + + G + P
Sbjct 216 AITNNDEFQQHLHFMQRAIGGVPSP 240
> CE07648
Length=392
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 65/206 (31%), Positives = 95/206 (46%), Gaps = 41/206 (19%)
Query 4 CFSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEY-VSRFGVEFDFVDFRDPSQV 62
FSSG+AAT ++ L GDH + +YGGT R V G+E D VD D +
Sbjct 75 VFSSGLAATSAIIN-LLKYGDHIVCSDDVYGGTQRYIRRVAVPNHGLEVDSVDLTDVQNL 133
Query 63 EKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVS 122
EKA K NTKMV E+P+NP L + D+ A+ Q + A P
Sbjct 134 EKAIKPNTKMVWFESPSNPLLKVVDIAAV--------------VQTAKKANP-------- 171
Query 123 DDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGG 182
I+ VVD+TF +P+ P GAD+++HS TKY +GH+ G
Sbjct 172 -----------------EIVVVVDNTFMSPYFQRPISLGADVVVHSITKYINGHSDVVMG 214
Query 183 AAISRFLKYHDKIAYTRNICGSIMDP 208
A I+ ++ + + + G++ P
Sbjct 215 AVITDNDEFQQHLFFMQLAVGAVPSP 240
> 7302582
Length=393
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 63/204 (30%), Positives = 91/204 (44%), Gaps = 40/204 (19%)
Query 5 FSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEK 64
FSSG+ AT VLT LS+GDH IM +YGGT R + +R G+ FVD ++
Sbjct 79 FSSGLGATTAVLTM-LSSGDHIIMGDDVYGGTNRLIRQVATRLGISATFVDPTKLDLIKS 137
Query 65 AFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDD 124
+ K TK+V E+P NP + + D+EAI ++
Sbjct 138 SIKPETKLVWIESPTNPLVKVADIEAIAQLVH---------------------------- 169
Query 125 ELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGAA 184
G RE I+ VD+TF T + P E GAD++ +S TKY +GH G
Sbjct 170 --------GVRE---DIVLAVDNTFLTSYFQRPLELGADLVCYSLTKYMNGHTDVVMGGI 218
Query 185 ISRFLKYHDKIAYTRNICGSIMDP 208
K + + + +N G + P
Sbjct 219 TMNSEKLYKSLKFLQNAVGIVPSP 242
> YAL012w
Length=394
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 62/207 (29%), Positives = 100/207 (48%), Gaps = 43/207 (20%)
Query 4 CFSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVD--FRDPSQ 61
FSSG A T T+L + L G H + +YGGT R + + GVE F + D Q
Sbjct 76 AFSSGSATTATILQS-LPQGSHAVSIGDVYGGTHRYFTKVANAHGVETSFTNDLLNDLPQ 134
Query 62 VEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPV 121
+ K+NTK+V ETP NPTL +TD++ +
Sbjct 135 L---IKENTKLVWIETPTNPTLKVTDIQKVA----------------------------- 162
Query 122 SDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTG 181
+L+K + G + ++ VVD+TF +P++ +P +GADI++HS TKY +GH+
Sbjct 163 ---DLIKKHAAG-----QDVILVVDNTFLSPYISNPLNFGADIVVHSATKYINGHSDVVL 214
Query 182 GAAISRFLKYHDKIAYTRNICGSIMDP 208
G + ++++ + +N G+I P
Sbjct 215 GVLATNNKPLYERLQFLQNAIGAIPSP 241
> HsM4503125
Length=405
Score = 96.3 bits (238), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 62/204 (30%), Positives = 90/204 (44%), Gaps = 41/204 (20%)
Query 5 FSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEK 64
F+SG+AAT+T+ T L AGD I +YGGT R + S FG++ FVD +E
Sbjct 87 FASGLAATVTI-THLLKAGDQIICMDDVYGGTNRYFRQVASEFGLKISFVDCSKIKLLEA 145
Query 65 AFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDD 124
A TK+V ETP NPT + D+E I+
Sbjct 146 AITPETKLVWIETPTNPTQKVIDIEGCAHIV----------------------------- 176
Query 125 ELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGAA 184
K I+ VVD+TF +P+ P GADI ++S TKY +GH+ G
Sbjct 177 -----------HKHGDIILVVDNTFMSPYFQRPLALGADISMYSATKYMNGHSDVVMGLV 225
Query 185 ISRFLKYHDKIAYTRNICGSIMDP 208
H+++ + +N G++ P
Sbjct 226 SVNCESLHNRLRFLQNSLGAVPSP 249
> Hs21361334
Length=405
Score = 96.3 bits (238), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 62/204 (30%), Positives = 90/204 (44%), Gaps = 41/204 (20%)
Query 5 FSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEK 64
F+SG+AAT+T+ T L AGD I +YGGT R + S FG++ FVD +E
Sbjct 87 FASGLAATVTI-THLLKAGDQIICMDDVYGGTNRYFRQVASEFGLKISFVDCSKIKLLEA 145
Query 65 AFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDD 124
A TK+V ETP NPT + D+E I+
Sbjct 146 AITPETKLVWIETPTNPTQKVIDIEGCAHIV----------------------------- 176
Query 125 ELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGAA 184
K I+ VVD+TF +P+ P GADI ++S TKY +GH+ G
Sbjct 177 -----------HKHGDIILVVDNTFMSPYFQRPLALGADISMYSATKYMNGHSDVVMGLV 225
Query 185 ISRFLKYHDKIAYTRNICGSIMDP 208
H+++ + +N G++ P
Sbjct 226 SVNCESLHNRLRFLQNSLGAVPSP 249
> CE17400
Length=444
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 65/213 (30%), Positives = 97/213 (45%), Gaps = 47/213 (22%)
Query 4 CFSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQ-- 61
++SG+AA V +LS+G H I+ +Y GT NE ++RFGVE VD
Sbjct 137 LYNSGLAAISAVFLEFLSSGAHMIVMNPIYSGTSSFINETLARFGVEITSVDVEKEKDFA 196
Query 62 --VEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAA 119
VEKA + NTKM+ E+ ANPT+ + D
Sbjct 197 GAVEKAIRPNTKMIYFESIANPTMAVPD-------------------------------- 224
Query 120 PVSDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGH-NI 178
+L TI + K IL +D+TF++P+ + P + GADI LHS +KY GH ++
Sbjct 225 ------ILGTIEVAKKYK---ILTCLDATFSSPYNIQPLKLGADISLHSCSKYIGGHTDV 275
Query 179 TTGGAAISRFLKYHDKIAYTRNICGSIMDPQTA 211
G +S + + K+ + GS + P A
Sbjct 276 IAGVVTVSSYDNW-KKLKLQQLTTGSSLSPYDA 307
> At3g57050
Length=464
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 52/207 (25%), Positives = 87/207 (42%), Gaps = 42/207 (20%)
Query 2 AACFSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQ 61
A CF+SGMAA ++ +T + G+ + +YGG+ R ++ V R GV V+ +
Sbjct 151 AFCFTSGMAA-LSAVTHLIKNGEEIVAGDDVYGGSDRLLSQVVPRSGVVVKRVNTTKLDE 209
Query 62 VEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPV 121
V A TK+V E+P NP ++D+ I +
Sbjct 210 VAAAIGPQTKLVWLESPTNPRQQISDIRKISEM--------------------------- 242
Query 122 SDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTG 181
+ L +VD++ +P + P E GADI++HS TK+ GH+
Sbjct 243 --------------AHAQGALVLVDNSIMSPVLSRPLELGADIVMHSATKFIAGHSDVMA 288
Query 182 GAAISRFLKYHDKIAYTRNICGSIMDP 208
G + K ++ + +N GS + P
Sbjct 289 GVLAVKGEKLAKEVYFLQNSEGSGLAP 315
> YLR303w
Length=444
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 51/168 (30%), Positives = 67/168 (39%), Gaps = 42/168 (25%)
Query 6 SSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEKA 65
SSG AA + GD+ + LYGGT+ RFG+E FV+ +P + EK
Sbjct 84 SSGQAAQTLAIQGLAHTGDNIVSTSYLYGGTYNQFKISFKRFGIEARFVEGDNPEEFEKV 143
Query 66 FKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDDE 125
F + TK V ET NP ++ D E I I K
Sbjct 144 FDERTKAVYLETIGNPKYNVPDFEKIVAIAHK---------------------------- 175
Query 126 LLKTISGGNREKERSILFVVDSTF-ATPHVLHPFEYGADIILHSTTKY 172
I VVD+TF A + P +YGADI+ HS TK+
Sbjct 176 -------------HGIPVVVDNTFGAGGYFCQPIKYGADIVTHSATKW 210
> At1g64660
Length=441
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 54/184 (29%), Positives = 81/184 (44%), Gaps = 44/184 (23%)
Query 2 AACFSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSR-FGVEFDFVDFRDPS 60
A C SSGM+A +V+ S+G H + LYGGT + ++ R + FVD D
Sbjct 119 AYCTSSGMSAISSVMLQLCSSGGHVVAASTLYGGTHALLSHFLPRTCNITTSFVDITDHG 178
Query 61 QVEKAF-KDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAA 119
V A + T+++ E+ ANPTL + D+ EL R+ +
Sbjct 179 AVANAIVEGRTQVLYFESVANPTLTVADI---------PELSRMAH-------------- 215
Query 120 PVSDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNIT 179
E+ + VVD+TFA P VL P + GAD+++HS +K+ G
Sbjct 216 ------------------EKGVTVVVDNTFA-PMVLSPAKLGADVVVHSISKFISGGADI 256
Query 180 TGGA 183
GA
Sbjct 257 IAGA 260
> At3g01120
Length=563
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 52/208 (25%), Positives = 78/208 (37%), Gaps = 43/208 (20%)
Query 5 FSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEK 64
+SGM A+ +L A + AG H + Y T ++ + G+ +D D + +E
Sbjct 253 MASGMCASTVMLLALVPAGGHIVTTTDCYRKTRIFMENFLPKLGITVTVIDPADIAGLEA 312
Query 65 AFKD-NTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSD 123
A + + E+P NP L D+E + I K
Sbjct 313 AVNEFKVSLFFTESPTNPFLRCVDIELVSKICHK-------------------------- 346
Query 124 DELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGA 183
R L +D TFATP GAD+++HS TKY GHN G
Sbjct 347 ---------------RGTLVCIDGTFATPLNQKALALGADLVVHSATKYIGGHNDVLAG- 390
Query 184 AISRFLKYHDKIAYTRNICGSIMDPQTA 211
I LK +I ++ G ++P A
Sbjct 391 CICGSLKLVSEIRNLHHVLGGTLNPNAA 418
> YFR055w
Length=340
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/172 (25%), Positives = 73/172 (42%), Gaps = 41/172 (23%)
Query 6 SSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEKA 65
+SG+ + L A AGDH +M +Y T + +++FGVE D+ D +EK
Sbjct 84 ASGLGSISLALLALSKAGDHILMTDSVYVPTRMLCDGLLAKFGVETDYYDPSIGKDIEKL 143
Query 66 FKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDDE 125
K NT ++ E+P + T+ + D+ A+ + +AKK
Sbjct 144 VKPNTTVIFLESPGSGTMEVQDIPAL-VSVAKK--------------------------- 175
Query 126 LLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHN 177
I ++D+T+ATP +G DI + + TKY GH+
Sbjct 176 -------------HGIKTILDNTWATPLFFDAHAHGIDISVEAGTKYLGGHS 214
> SPBC428.11
Length=429
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 47/172 (27%), Positives = 70/172 (40%), Gaps = 42/172 (24%)
Query 2 AACFSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQ 61
A SSG +A LT AGD+ + LYGGT+ + R G+ FV+ DP+
Sbjct 81 AIATSSGTSALFMALTTLAKAGDNIVSTSYLYGGTYNLFKVTLPRLGITTKFVNGDDPND 140
Query 62 VEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPV 121
+ +NTK V E+ NP ++ D E I + AA V
Sbjct 141 LAAQIDENTKAVYVESIGNPMYNVPDFERIAEV---------------------AHAAGV 179
Query 122 SDDELLKTISGGNREKERSILFVVDSTF-ATPHVLHPFEYGADIILHSTTKY 172
+VD+TF +++ P ++GADI+ HS TK+
Sbjct 180 P--------------------LMVDNTFGGGGYLVRPIDHGADIVTHSATKW 211
> At1g33320
Length=412
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 46/208 (22%), Positives = 78/208 (37%), Gaps = 43/208 (20%)
Query 5 FSSGMAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRFGVEFDFVDFRDPSQVEK 64
+SGM A+ +L A + H + Y T ++++ G+ F+D D + ++
Sbjct 103 MASGMYASNVMLLALVPTNGHIVATKDCYKETRIFMENFLTKLGITVTFIDSDDIAGLQT 162
Query 65 AFKDN-TKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSD 123
++ + E+P NP L D++ + I +
Sbjct 163 LVNNHEVSLFFTESPTNPFLRCVDIKLVSKICHR-------------------------- 196
Query 124 DELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGA 183
R L +D+T ATP GAD++ HS TKY GHN G
Sbjct 197 ---------------RGTLVCIDATIATPINQKTLALGADLVHHSATKYIGGHNDFLAG- 240
Query 184 AISRFLKYHDKIAYTRNICGSIMDPQTA 211
+IS ++ KI + G ++P A
Sbjct 241 SISGSMELVSKIRNLHKLLGGTLNPNAA 268
> YHR112c
Length=378
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 43/78 (55%), Gaps = 7/78 (8%)
Query 131 SGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGA------A 184
S R + L +VDSTFA+P + + + +GADI+L+S TKYF GH+ G A
Sbjct 158 SLARRAHAKGALLIVDSTFASPPLQYAWNFGADIVLYSATKYFGGHSDLLSGVLVVKEEA 217
Query 185 ISRFLKYHDKIAYTRNIC 202
SR LK D+I N+
Sbjct 218 TSRQLK-DDRIYLGTNVA 234
> SPAC23A1.14c
Length=398
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 137 KERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITTGGAAISRFLKYHDKIA 196
K++ + VVDSTFA P + GAD ++HS TKY GH+ G S+ +
Sbjct 178 KKKGAILVVDSTFAPPPIQDALVLGADYVVHSATKYLAGHSDVLAGVTASKDRSKILDLK 237
Query 197 YTRNICGSIMDPQTA 211
R G+I+ PQ A
Sbjct 238 ADRAYLGTILHPQQA 252
> YGL184c
Length=465
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 57/218 (26%), Positives = 81/218 (37%), Gaps = 59/218 (27%)
Query 6 SSGMAATITVL------------TAYLSAGDHCIMPICLYGGTFRAANEYVSR---FGVE 50
SSGM A +L T + AGD LYGGT R N + + V
Sbjct 77 SSGMTALDVILRGLVLLNGTDNHTPTIIAGDD------LYGGTQRLLNFFKQQSHAVSVH 130
Query 51 FDFVDFRDPSQVEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTE 110
D DF V ++ D VL E+P NP + D+ I +
Sbjct 131 VDTSDFEKFKTVFQSL-DKVDCVLLESPTNPLCKVVDIPRILRFVK-------------- 175
Query 111 NATPSTDAAPVSDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTT 170
S D V D+ ++ SG N P L+P G D++ S T
Sbjct 176 --CISPDTTVVVDNTMM---SGLN---------------CNPLQLNP---GCDVVYESAT 212
Query 171 KYFDGHNITTGGAAISRFLKYHDKIAYTRNICGSIMDP 208
KY +GH+ GG IS+ + K+ + N G+ + P
Sbjct 213 KYLNGHHDLMGGVIISKTPEIASKLYFVINSTGAGLSP 250
> YLL058w
Length=575
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 39/67 (58%), Gaps = 5/67 (7%)
Query 30 ICLYGGTFRAANEYVSRFGV--EFDFVDFRDPSQVEKAFKDNTKMVLA---ETPANPTLH 84
+ ++G F+ +++FG F F + RD +++K + + + +LA ETP+NP L+
Sbjct 273 VGIFGFPFKDTQVIMTKFGKCKFFGFGNSRDVVELQKFLETSKQRILAVFVETPSNPLLN 332
Query 85 LTDLEAI 91
+ DL+ +
Sbjct 333 MPDLKKL 339
> Hs4504139
Length=877
Score = 33.9 bits (76), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 5/61 (8%)
Query 121 VSDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNITT 180
V D L I N E+E I+FVV++ +A H LH + + +TTK D I
Sbjct 370 VCDKHL--AIDSSNYEQESKIMFVVNAVYAMAHALHKMQ---RTLCPNTTKLCDAMKILD 424
Query 181 G 181
G
Sbjct 425 G 425
> Hs20070337
Length=250
Score = 32.7 bits (73), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 39/86 (45%), Gaps = 16/86 (18%)
Query 33 YGGTFRAANEYVSRFGVEF-------DFVD-FRDPSQV-EKAFKDNTKMVLAETPANPTL 83
+G TFRA N R EF FVD +D QV E+ FK T MVL++ P
Sbjct 11 FGPTFRAENSQSRRHLAEFYMIEAEISFVDSLQDLMQVIEELFKATTMMVLSKCPE---- 66
Query 84 HLTDLEAIDMIIAKKELERLRNKQKT 109
D+E IA + +RL + K
Sbjct 67 ---DVELCHKFIAPGQKDRLEHMLKN 89
> YMR189w
Length=1034
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 69/148 (46%), Gaps = 14/148 (9%)
Query 62 VEKAFKDNTKMVLAETPANP-TLHLTDLEAIDMIIAKKELERLRNKQKTENAT--PSTDA 118
++K TK VL T A P + + +++ D+ KK ++ L+N + P+TD
Sbjct 231 IDKKLHQQTKSVL-HTRAKPFNIEIIEVDCSDI---KKAVDVLKNPDVSGCLVQYPATDG 286
Query 119 APVSDDELLKTISGGNREKERSILFVVDSTFATPHVLHPFEYGADIILHSTTKYFDGHNI 178
+ + D + K +S +S+L V A + P YGADI+L S+ ++ G +
Sbjct 287 SILPPDSM-KQLSDA-LHSHKSLLSVASDLMALTLLKPPAHYGADIVLGSSQRF--GVPM 342
Query 179 TTGGAAISRFLKYHDKIAYTRNICGSIM 206
GG + F DK+ R I G I+
Sbjct 343 GYGGPH-AAFFAVIDKL--NRKIPGRIV 367
> CE10298
Length=464
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 26/101 (25%), Positives = 46/101 (45%), Gaps = 5/101 (4%)
Query 1 DAACFSSGMAATITV-LTAYLSAGDHCIMP---ICLYGGTFRAANEYVSRFGVEFDFVDF 56
D +SG + + + + A +AG++ ++P LY R N + ++ D
Sbjct 139 DDVVLASGCSHALQMAIEAVANAGENILVPHPGFPLYSTLCRPHNIVDKPYKIDMTGEDV 198
Query 57 R-DPSQVEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIA 96
R D S + DNTK ++ P NPT + E ++ I+A
Sbjct 199 RIDLSYMATIIDDNTKAIIVNNPGNPTGGVFTKEHLEEILA 239
> 7299520
Length=417
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 9/60 (15%)
Query 58 DPSQVEKAFKDNTKMVLAETPANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTD 117
D ++ E F TKM++ TP NP I + +KELER+ + N +D
Sbjct 162 DDAEFESLFNSKTKMIILNTPHNP---------IGKVFNRKELERIAELCRKWNVLCVSD 212
> Hs21071039
Length=507
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 18/83 (21%), Positives = 39/83 (46%), Gaps = 2/83 (2%)
Query 9 MAATITVLTAYLSAGDHCIMPICLYGGTFRAANEYVSRF-GVEFDFVDFRDPSQVEKAFK 67
MA + +L + + + H ++P +Y E ++ + + D ++R+ S+VEK
Sbjct 273 MADLVALLGSLVDSSGHILVP-GIYDEVVPLTEEEINTYKAIHLDLEEYRNSSRVEKFLF 331
Query 68 DNTKMVLAETPANPTLHLTDLEA 90
D + +L P+L + +E
Sbjct 332 DTKEEILMHLWRYPSLSIHGIEG 354
> HsM9625014
Length=175
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 47 FGVEFDFVDFR-DPSQVEKAFKDNTKMVLAETPANPTLHLTDLEAIDMI 94
+G + D+ DP ++E F TK ++ TP NP + + E + +I
Sbjct 20 YGKRWSSSDWTLDPQELESKFNPKTKAIILNTPHNPLGKVYNREELQVI 68
> At1g68790
Length=1085
Score = 30.0 bits (66), Expect = 3.9, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query 99 ELERLRNKQKTENATPSTDAAPVSDDELLKTISGGNREKERSILFVVDST 148
E R R +++T T S AA SD E + +I+ G R K+R I V T
Sbjct 887 ETPRKRQREQTSRITESEQAAGDSD-EGVDSITTGGRRKKRQIAVPVSQT 935
> CE27123
Length=1596
Score = 28.9 bits (63), Expect = 7.3, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 43/93 (46%), Gaps = 11/93 (11%)
Query 70 TKMVLAETP---ANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDDEL 126
T + AETP PT D E DM E++R+ ++ K + A PST + ++
Sbjct 268 TAPIAAETPKTKVTPTSSEDDWEKADM-----EVQRVEDENKRQKAVPSTAVSEKPEESR 322
Query 127 LKTISGGNREKERSILFVVDSTFATPHVL-HPF 158
+ GG+ ++ + + + P +L HPF
Sbjct 323 ESSSLGGSWSQQDT--EPSERSSVEPEILEHPF 353
> CE04378
Length=1592
Score = 28.9 bits (63), Expect = 7.3, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 43/93 (46%), Gaps = 11/93 (11%)
Query 70 TKMVLAETP---ANPTLHLTDLEAIDMIIAKKELERLRNKQKTENATPSTDAAPVSDDEL 126
T + AETP PT D E DM E++R+ ++ K + A PST + ++
Sbjct 268 TAPIAAETPKTKVTPTSSEDDWEKADM-----EVQRVEDENKRQKAVPSTAVSEKPEESR 322
Query 127 LKTISGGNREKERSILFVVDSTFATPHVL-HPF 158
+ GG+ ++ + + + P +L HPF
Sbjct 323 ESSSLGGSWSQQDT--EPSERSSVEPEILEHPF 353
Lambda K H
0.319 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3825978242
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40