bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0341_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
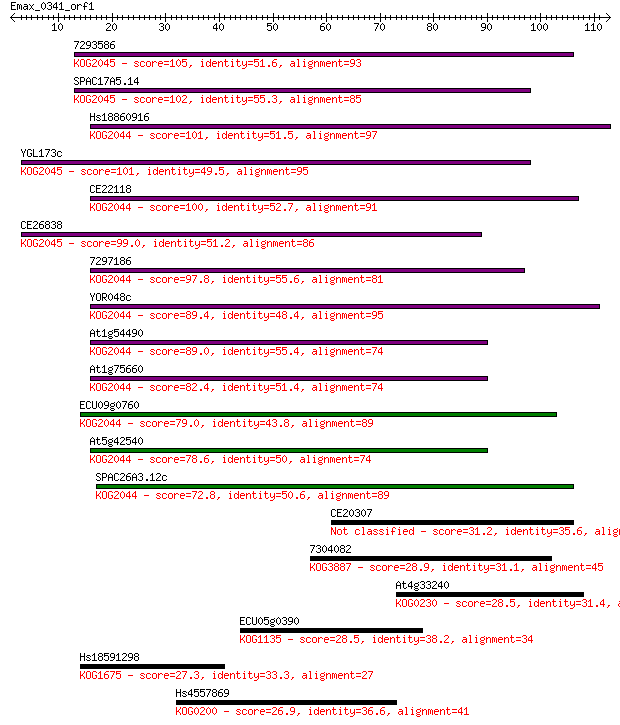
7293586 105 3e-23
SPAC17A5.14 102 2e-22
Hs18860916 101 3e-22
YGL173c 101 3e-22
CE22118 100 9e-22
CE26838 99.0 2e-21
7297186 97.8 4e-21
YOR048c 89.4 1e-18
At1g54490 89.0 2e-18
At1g75660 82.4 2e-16
ECU09g0760 79.0 2e-15
At5g42540 78.6 2e-15
SPAC26A3.12c 72.8 1e-13
CE20307 31.2 0.56
7304082 28.9 2.3
At4g33240 28.5 3.0
ECU05g0390 28.5 3.0
Hs18591298 27.3 6.2
Hs4557869 26.9 9.9
> 7293586
Length=1612
Score = 105 bits (261), Expect = 3e-23, Method: Composition-based stats.
Identities = 48/94 (51%), Positives = 68/94 (72%), Gaps = 2/94 (2%)
Query 13 IPTFDNLYLDLSGILHNCSHGNSGGM-LHANEDLMWQSVFAALDLVISTISPRKLLYLAA 71
IP FDNLYLD++GI+HNCSH + + H E+ ++Q +F +D + I P++L +L+
Sbjct 26 IPEFDNLYLDMNGIVHNCSHPDDNNIHFHLEEEQIFQEIFNYVDKLFYLIKPQRLFFLSV 85
Query 72 DGVAPRAKMNQQRARRYRAAKSAKEAAEAQAKQQ 105
DGVAPRAKMNQQR+RR+R A+ A E EA+A Q+
Sbjct 86 DGVAPRAKMNQQRSRRFRTAREA-EQQEAKAAQR 118
> SPAC17A5.14
Length=1328
Score = 102 bits (253), Expect = 2e-22, Method: Composition-based stats.
Identities = 47/89 (52%), Positives = 64/89 (71%), Gaps = 7/89 (7%)
Query 13 IPTFDNLYLDLSGILHNCSHGNSGGMLHAN----EDLMWQSVFAALDLVISTISPRKLLY 68
IP FDNLYLD++GILHNC+H N H++ E+ M+ ++F ++ + I P+KLLY
Sbjct 26 IPEFDNLYLDMNGILHNCTHKNDD---HSSPPLPEEEMYIAIFNYIEHLFEKIKPKKLLY 82
Query 69 LAADGVAPRAKMNQQRARRYRAAKSAKEA 97
+A DG APRAKMNQQR+RR+R AK A +A
Sbjct 83 MAVDGCAPRAKMNQQRSRRFRTAKDAHDA 111
> Hs18860916
Length=950
Score = 101 bits (252), Expect = 3e-22, Method: Composition-based stats.
Identities = 50/97 (51%), Positives = 66/97 (68%), Gaps = 4/97 (4%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M ++F +D + S + PR+LLY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDKPA-PKNEDEMMVAIFEYIDRLFSIVRPRRLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTEWL 112
PRAKMNQQR+RR+RA +KE EA ++QR E L
Sbjct 107 PRAKMNQQRSRRFRA---SKEGMEAAVEKQRVREEIL 140
> YGL173c
Length=1528
Score = 101 bits (252), Expect = 3e-22, Method: Composition-based stats.
Identities = 47/96 (48%), Positives = 66/96 (68%), Gaps = 1/96 (1%)
Query 3 VINEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGML-HANEDLMWQSVFAALDLVISTI 61
+I + I + IP FDNLYLD++ ILHNC+HGN + E+ ++ + +D + TI
Sbjct 16 MILQLIEGTQIPEFDNLYLDMNSILHNCTHGNDDDVTKRLTEEEVFAKICTYIDHLFQTI 75
Query 62 SPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEA 97
P+K+ Y+A DGVAPRAKMNQQRARR+R A A++A
Sbjct 76 KPKKIFYMAIDGVAPRAKMNQQRARRFRTAMDAEKA 111
> CE22118
Length=892
Score = 100 bits (248), Expect = 9e-22, Method: Composition-based stats.
Identities = 48/91 (52%), Positives = 64/91 (70%), Gaps = 2/91 (2%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M+ +F +D + S + PR+LLY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDRPA-PKNEDEMFALIFEYIDRIYSIVRPRRLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQR 106
PRAKMNQQR+RR+RA+K E EA ++QR
Sbjct 107 PRAKMNQQRSRRFRASKEMAE-KEASIEEQR 136
> CE26838
Length=1209
Score = 99.0 bits (245), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 44/87 (50%), Positives = 63/87 (72%), Gaps = 1/87 (1%)
Query 3 VINEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGM-LHANEDLMWQSVFAALDLVISTI 61
++E I S IP FDNLYLD++GI+HNCSH N + ED ++ ++FA ++ + + I
Sbjct 16 CLSEVINESQIPEFDNLYLDMNGIIHNCSHPNDDDVTFRITEDEIFVNIFAYIENLYNLI 75
Query 62 SPRKLLYLAADGVAPRAKMNQQRARRY 88
P+K+ ++A DGVAPRAKMNQQRARR+
Sbjct 76 RPQKVFFMAVDGVAPRAKMNQQRARRF 102
> 7297186
Length=885
Score = 97.8 bits (242), Expect = 4e-21, Method: Composition-based stats.
Identities = 45/81 (55%), Positives = 57/81 (70%), Gaps = 1/81 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M ++F +D + + PRKLLY+A DGVA
Sbjct 49 FDNLYLDMNGIIHPCTHPEDKPA-PKNEDEMMVAIFECIDRLFGIVRPRKLLYMAIDGVA 107
Query 76 PRAKMNQQRARRYRAAKSAKE 96
PRAKMNQQR+RR+RAAK E
Sbjct 108 PRAKMNQQRSRRFRAAKETTE 128
> YOR048c
Length=1006
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 46/95 (48%), Positives = 62/95 (65%), Gaps = 1/95 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
DNLYLD++GI+H CSH + ED M +VF + V++ PRK+L +A DGVA
Sbjct 47 LDNLYLDMNGIVHPCSHPENKPPPET-EDEMLLAVFEYTNRVLNMARPRKVLVMAVDGVA 105
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTE 110
PRAKMNQQRARR+R+A+ A+ EA+ + R E
Sbjct 106 PRAKMNQQRARRFRSARDAQIENEAREEIMRQREE 140
> At1g54490
Length=947
Score = 89.0 bits (219), Expect = 2e-18, Method: Composition-based stats.
Identities = 41/74 (55%), Positives = 55/74 (74%), Gaps = 1/74 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C H G A D +++S+F +D + + + PRK+LYLA DGVA
Sbjct 49 FDNLYLDMNGIIHPCFH-PEGKPAPATYDDVFKSMFEYIDHLFTLVRPRKILYLAIDGVA 107
Query 76 PRAKMNQQRARRYR 89
PRAKMNQQR+RR+R
Sbjct 108 PRAKMNQQRSRRFR 121
> At1g75660
Length=985
Score = 82.4 bits (202), Expect = 2e-16, Method: Composition-based stats.
Identities = 38/74 (51%), Positives = 52/74 (70%), Gaps = 1/74 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
+DNLYLD++GI+H C H E++ +Q +F +D + + PRKLLY+A DGVA
Sbjct 48 YDNLYLDMNGIIHPCFHPEDRPSPTTFEEV-FQCMFDYIDRLFVMVRPRKLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYR 89
PRAKMNQQR+RR+R
Sbjct 107 PRAKMNQQRSRRFR 120
> ECU09g0760
Length=655
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 39/89 (43%), Positives = 54/89 (60%), Gaps = 1/89 (1%)
Query 14 PTFDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADG 73
P+ D LYLD + I+H C G DL +++V +D ++ PRKLLY+A DG
Sbjct 25 PSVDALYLDFNAIIHTCIKPGLGSEEEIERDL-YKTVGDFIDHILLKTRPRKLLYIAVDG 83
Query 74 VAPRAKMNQQRARRYRAAKSAKEAAEAQA 102
VAPRAK++ QRARRY++A KE + A
Sbjct 84 VAPRAKLSHQRARRYKSALGKKEGSAVIA 112
> At5g42540
Length=1012
Score = 78.6 bits (192), Expect = 2e-15, Method: Composition-based stats.
Identities = 37/74 (50%), Positives = 51/74 (68%), Gaps = 1/74 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
+DNLYLD++GI+H C H ++ +Q +F +D + + PRKLL++A DGVA
Sbjct 50 YDNLYLDMNGIIHPCFHPEDKPSPTTFTEV-FQCMFDYIDRLFVMVRPRKLLFMAIDGVA 108
Query 76 PRAKMNQQRARRYR 89
PRAKMNQQRARR+R
Sbjct 109 PRAKMNQQRARRFR 122
> SPAC26A3.12c
Length=991
Score = 72.8 bits (177), Expect = 1e-13, Method: Composition-based stats.
Identities = 45/95 (47%), Positives = 62/95 (65%), Gaps = 7/95 (7%)
Query 17 DNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVAP 76
DNLYLD++GI+H CSH ED M +VF D +++ + PR+LL++A DGVAP
Sbjct 50 DNLYLDMNGIVHPCSHPEDRPAPET-EDEMMVAVFEYTDRILAMVRPRQLLFIAIDGVAP 108
Query 77 RAKMNQQRARRYRAAKSAKEAAE------AQAKQQ 105
RAKMNQQR+RR+R+++ A E +AKQQ
Sbjct 109 RAKMNQQRSRRFRSSREAALKEEELQAFIEEAKQQ 143
> CE20307
Length=485
Score = 31.2 bits (69), Expect = 0.56, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 3/48 (6%)
Query 61 ISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKE---AAEAQAKQQ 105
I PRK++ + P+AKM ++R R + K +E A E K+Q
Sbjct 227 IPPRKIIEKPVEKAPPKAKMTRKRMMRAKHEKRLREEQIALEESKKEQ 274
> 7304082
Length=387
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 57 VISTISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEAAEAQ 101
+I I+ R +L+ DG++ +KM QR R++ EA Q
Sbjct 166 LIVLITQRHMLFSQVDGISDDSKMESQRDIHQRSSDDLNEAGLDQ 210
> At4g33240
Length=1757
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 11/35 (31%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 73 GVAPRAKMNQQRARRYRAAKSAKEAAEAQAKQQRA 107
G +PR KMN + +RR + ++++ + AK +R+
Sbjct 145 GPSPRPKMNPRASRRVSSNMDSEKSEQQNAKSRRS 179
> ECU05g0390
Length=639
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 44 DLMWQSVFAALDLVISTISPRKLLYLAADGVAPR 77
DL+ S ++ ++I T+SPRKL+ + DG R
Sbjct 466 DLVGISDLSSCKMIIETLSPRKLVCVGEDGDTER 499
> Hs18591298
Length=753
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 9/27 (33%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 14 PTFDNLYLDLSGILHNCSHGNSGGMLH 40
P + L ++LS ++H+C H G+ H
Sbjct 347 PRYSQLAMELSQVVHSCPHETQPGVYH 373
> Hs4557869
Length=1124
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 32 HGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAAD 72
HGN L + L FA + ST+S ++LL+ AAD
Sbjct 907 HGNLLDFLRKSRVLETDPAFAIANSTASTLSSQQLLHFAAD 947
Lambda K H
0.319 0.129 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1188972946
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40