bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
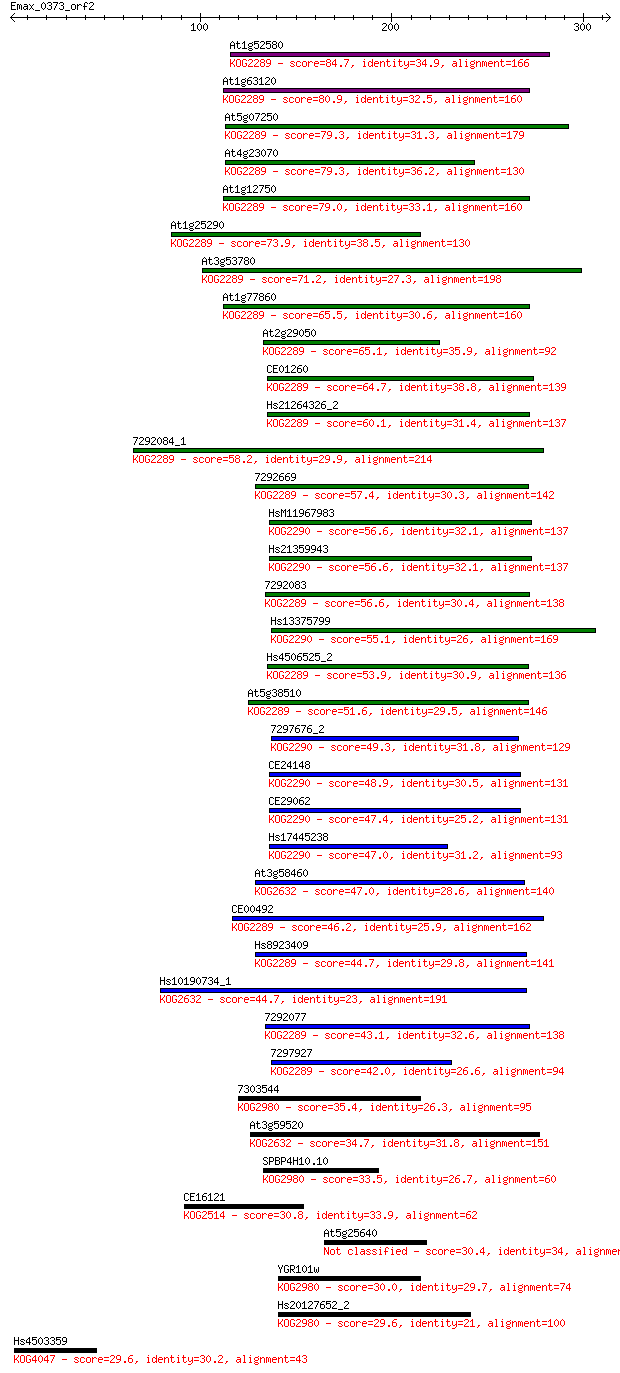
Query= Emax_0373_orf2
Length=313
Score E
Sequences producing significant alignments: (Bits) Value
At1g52580 84.7 2e-16
At1g63120 80.9 3e-15
At5g07250 79.3 9e-15
At4g23070 79.3 9e-15
At1g12750 79.0 1e-14
At1g25290 73.9 4e-13
At3g53780 71.2 3e-12
At1g77860 65.5 2e-10
At2g29050 65.1 2e-10
CE01260 64.7 2e-10
Hs21264326_2 60.1 6e-09
7292084_1 58.2 2e-08
7292669 57.4 4e-08
HsM11967983 56.6 6e-08
Hs21359943 56.6 6e-08
7292083 56.6 7e-08
Hs13375799 55.1 2e-07
Hs4506525_2 53.9 4e-07
At5g38510 51.6 2e-06
7297676_2 49.3 1e-05
CE24148 48.9 1e-05
CE29062 47.4 4e-05
Hs17445238 47.0 5e-05
At3g58460 47.0 6e-05
CE00492 46.2 9e-05
Hs8923409 44.7 2e-04
Hs10190734_1 44.7 3e-04
7292077 43.1 8e-04
7297927 42.0 0.001
7303544 35.4 0.16
At3g59520 34.7 0.27
SPBP4H10.10 33.5 0.68
CE16121 30.8 3.6
At5g25640 30.4 4.8
YGR101w 30.0 6.7
Hs20127652_2 29.6 8.0
Hs4503359 29.6 8.3
> At1g52580
Length=309
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 58/173 (33%), Positives = 86/173 (49%), Gaps = 7/173 (4%)
Query 116 SPSISSLITFGASIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTR 175
PSI +L GA + G+ WRLI ++LH LHL NM + IG +E+++G
Sbjct 84 GPSIPTLRKLGALERRLVEEGERWRLISCIWLHGGFLHLMANMISLMCIGMRLEQEFGFM 143
Query 176 RICVAYLVCALLGNLVSAATFF-CNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVV 234
RI Y++ L G+LVS T + VG+S A+FGL+G L+EL+ W E++ +
Sbjct 144 RIGALYVISGLGGSLVSCLTDSQGERVSVGASGALFGLLGAMLSELITNWTIYENKCTAL 203
Query 235 LQLVLFGLLFVFLSIGSATDVVGHLAGMAAGLSFGLV------YNSSNPQGSP 281
+ L+L +L + + D H G AG G V Y NP+ P
Sbjct 204 MTLILIIVLNLSVGFLPRVDNSAHFGGFLAGFFLGFVLLLRPQYGYVNPKYIP 256
> At1g63120
Length=317
Score = 80.9 bits (198), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 52/162 (32%), Positives = 85/162 (52%), Gaps = 3/162 (1%)
Query 112 DNPT-SPSISSLITFGA-SIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIE 169
+NP PS S+L GA + Q WRL+ ++LH I+HL NM I IG +E
Sbjct 82 ENPLFGPSSSTLEKMGALEWRKVVHEHQGWRLLSCMWLHAGIIHLLTNMLSLIFIGIRLE 141
Query 170 EKYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIED 229
+++G R+ + YL+ L G+++S + F ++ VG+S A+FGL+G L+ELL W +
Sbjct 142 QQFGFIRVGLIYLISGLGGSILS-SLFLQESISVGASGALFGLLGAMLSELLTNWTIYAN 200
Query 230 RRPVVLQLVLFGLLFVFLSIGSATDVVGHLAGMAAGLSFGLV 271
+ ++ L+ + + L + D H+ G G G V
Sbjct 201 KAAALITLLFIIAINLALGMLPRVDNFAHIGGFLTGFCLGFV 242
> At5g07250
Length=346
Score = 79.3 bits (194), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 56/191 (29%), Positives = 92/191 (48%), Gaps = 13/191 (6%)
Query 113 NPT-SPSISSLITFGA-SIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEE 170
NP PS +L GA + + + WRL+ ++LH ++HL NM + IG +E+
Sbjct 107 NPLFGPSSHTLEKLGALEWSKVVEKKEGWRLLTCIWLHAGVIHLGANMLSLVFIGIRLEQ 166
Query 171 KYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDR 230
++G RI V YL+ + G+++S+ F N++ VG+S A+FGL+G L+EL W ++
Sbjct 167 QFGFVRIGVIYLLSGIGGSVLSS-LFIRNSISVGASGALFGLLGSMLSELFTNWTIYSNK 225
Query 231 RPVVLQLVLFGLLFVFLSIGSATDVVGHLAGMAAGLSFGLVY----------NSSNPQGS 280
+L L+ L+ + + I D H+ G G G + PQG+
Sbjct 226 IAALLTLLFVILINLAIGILPHVDNFAHVGGFVTGFLLGFILLARPQFKWLAREHMPQGT 285
Query 281 PHVDVYTRISY 291
P Y Y
Sbjct 286 PLRYKYKTYQY 296
> At4g23070
Length=313
Score = 79.3 bits (194), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 81/133 (60%), Gaps = 4/133 (3%)
Query 113 NPT-SPSISSLITFGA-SIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEE 170
NP PS S+L GA + + + Q+WRL+ ++LH ++HL NM IG +E+
Sbjct 77 NPLLGPSSSTLEKMGALAWGKIVHKRQVWRLLTCMWLHAGVIHLLANMCCVAYIGVRLEQ 136
Query 171 KYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDR 230
++G R+ YLV G+++S F +A+ VG+S+A+FGL+G L+ELL+ W + +++
Sbjct 137 QFGFVRVGTIYLVSGFCGSILS-CLFLEDAISVGASSALFGLLGAMLSELLINWTTYDNK 195
Query 231 R-PVVLQLVLFGL 242
+V+ LV+ G+
Sbjct 196 GVAIVMLLVIVGV 208
> At1g12750
Length=307
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/164 (32%), Positives = 88/164 (53%), Gaps = 7/164 (4%)
Query 112 DNP-TSPSISSLITFGA-SIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIE 169
+NP PS S+L GA ++ + WRLI ++LH I+HL +NMF I G +E
Sbjct 69 ENPFLGPSSSTLEKLGALDWKKVVQGNEKWRLITAMWLHAGIIHLVMNMFDVIIFGIRLE 128
Query 170 EKYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIED 229
+++G RI + YL+ G+++S A F ++ VG+S A+ GL+G L+ELL W +
Sbjct 129 QQFGFIRIGLIYLISGFGGSILS-ALFLQKSISVGASGALLGLMGAMLSELLTNWTIYKS 187
Query 230 RRPVVLQLVLFGLLFVFLSIG--SATDVVGHLAGMAAGLSFGLV 271
+ +L + ++ + L+IG D H+ G+ G G +
Sbjct 188 KLCALLSFLF--IIAINLAIGLLPWVDNFAHIGGLLTGFCLGFI 229
> At1g25290
Length=369
Score = 73.9 bits (180), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 50/135 (37%), Positives = 70/135 (51%), Gaps = 16/135 (11%)
Query 85 NAKHCVFILAVIDLIVYIATVIYGQSPDNPTSPSISSLITFGASIPSAIRRGQIWRLICP 144
N + +L I++I+YIA + S ++T+GA I S I RGQ+WRL
Sbjct 151 NGRRWTNVLLAINVIMYIAQI-----------ASDGKVLTWGAKINSLIERGQLWRLATA 199
Query 145 VFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCA----LLGNLVSAATFFCN- 199
LH N +HL IN + IG + E G +R YL A +L L SA +++ N
Sbjct 200 SVLHANPMHLMINCYSLNSIGPTAESLGGPKRFLAVYLTSAVAKPILRVLGSAMSYWFNK 259
Query 200 ALKVGSSTAIFGLIG 214
A VG+S AIFGL+G
Sbjct 260 APSVGASGAIFGLVG 274
> At3g53780
Length=361
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 54/211 (25%), Positives = 100/211 (47%), Gaps = 19/211 (9%)
Query 101 YIATVIYGQSPDNPT-SPSISSLITFGA-SIPSAIRRGQIWRLICPVFLHLNILHLFINM 158
++ + + +NP PS +L T G + ++ + WRL+ +LH ++HL +NM
Sbjct 72 FLGRFSFQNTRENPLLGPSSLTLQTMGGLDVKKVVKGDEGWRLLSCNWLHGGVVHLLMNM 131
Query 159 FMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLA 218
+ IG +E RI + YL+ G+++SA F + + VG+S A+FGL+G L+
Sbjct 132 LTLLFIGIRME-----LRIGLLYLISGFGGSILSA-LFLRSNISVGASGAVFGLLGGMLS 185
Query 219 ELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSATDVVGHLAGMAAGL----------SF 268
E+ + W ++ ++ LVL + + L + D H+ G A G +
Sbjct 186 EIFINWTIYSNKVVTIVTLVLIVAVNLGLGVLPGVDNFAHIGGFATGFLLGFVLLIRPHY 245
Query 269 GLVYNSSNPQGSPH-VDVYTRISYAFNAVIV 298
G + + P PH +Y I + + +I+
Sbjct 246 GWINQRNGPGAKPHRFKIYQGILWTISLLIL 276
> At1g77860
Length=319
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 49/165 (29%), Positives = 77/165 (46%), Gaps = 9/165 (5%)
Query 112 DNPT-SPSISSLITFGA-SIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIE 169
+NP PS S+L G S + +IWR++ +LH + HLFIN+ I +G +E
Sbjct 94 ENPMLGPSASTLEHMGGLSWKALTENHEIWRILTSPWLHSGLFHLFINLGSLIFVGIYME 153
Query 170 EKYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIED 229
+++G RI V Y + ++G+L A F N + S A FGLIG L+ L W+
Sbjct 154 QQFGPLRIAVIYFLSGIMGSLF-AVLFVRNIPSISSGAAFFGLIGAMLSALAKNWNLYNS 212
Query 230 R---RPVVLQLVLFGLLFVFLSIGSATDVVGHLAGMAAGLSFGLV 271
+ ++ + L FL D ++ G +G G V
Sbjct 213 KISALAIIFTIFTVNFLIGFLPF---IDNFANIGGFISGFLLGFV 254
> At2g29050
Length=372
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 56/92 (60%), Gaps = 1/92 (1%)
Query 133 IRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVS 192
+ + ++WRL ++LH + H+ NM I IG +E+++G RI + Y++ G+L+S
Sbjct 150 VHKHEVWRLFTCIWLHAGVFHVLANMLSLIFIGIRLEQEFGFVRIGLLYMISGFGGSLLS 209
Query 193 AATFFCNALKVGSSTAIFGLIGIDLAELLVIW 224
+ F + VG+S A+FGL+G L+ELL W
Sbjct 210 -SLFNRAGISVGASGALFGLLGAMLSELLTNW 240
> CE01260
Length=419
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 54/153 (35%), Positives = 77/153 (50%), Gaps = 19/153 (12%)
Query 135 RGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAA 194
RG+ WR +FLH + HL N+ +Q+ +G +E + RI YL+ G+L+ A
Sbjct 226 RGEAWRFTSYMFLHAGLNHLLGNVIIQLLVGIPLEVAHKIWRIGPIYLLAVTSGSLLQYA 285
Query 195 TFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSA-- 252
N+L VG+S ++ LI +A +++ WH + R V LVLF +F+FL G A
Sbjct 286 I-DPNSLLVGASAGVYALIFAHVANVILNWHEMPLRWIRV--LVLF--VFIFLDFGGAIH 340
Query 253 -------TDVVGHL---AGMAAGLSFGLV--YN 273
D V HL AG GL FG V YN
Sbjct 341 RRFYTNDCDSVSHLAHIAGAVTGLFFGYVVLYN 373
> Hs21264326_2
Length=302
Score = 60.1 bits (144), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 43/153 (28%), Positives = 76/153 (49%), Gaps = 20/153 (13%)
Query 135 RGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNL-VSA 193
R Q+WR + +F+H I HL +N+ +Q+ +G +E +G RI + Y+ + G+L VS
Sbjct 106 RAQVWRYLTYIFMHAGIEHLGLNVVLQLLVGVPLEMVHGATRIGLVYVAGVVAGSLAVSV 165
Query 194 ATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSAT 253
A A VGSS ++ L+ LA +++ W ++ + ++ V L+ + + G A
Sbjct 166 ADM--TAPVVGSSGGVYALVSAHLANIVMNWSGMKCQFKLLRMAV--ALICMSMEFGRAV 221
Query 254 ---------------DVVGHLAGMAAGLSFGLV 271
V HL G+A G++ G+V
Sbjct 222 WLRFHPSAYPPCPHPSFVAHLGGVAVGITLGVV 254
> 7292084_1
Length=356
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 64/241 (26%), Positives = 106/241 (43%), Gaps = 41/241 (17%)
Query 65 SRFRLWFSGRSLVECLLPKFNAKHCVFILAVIDL--IVYIATVIYGQSPDNPTSPSISSL 122
R R +++ R C P F + ++ +++L VY + V +P P PS S
Sbjct 51 ERDRKYYADR--YTCCPPPF----FIILVTLVELGFFVYHSVVTGEAAPRGPI-PSDSMF 103
Query 123 ITFGASIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYL 182
I +R +IWR + + LH LHL N+ +Q+ G +E +G+ RI Y
Sbjct 104 IY------RPDKRHEIWRFLFYMVLHAGWLHLGFNVAVQLVFGLPLEMVHGSTRIACIYF 157
Query 183 VCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLF-G 241
L G+L + F + VG+S ++ L+ LA +L+ +H + +L +++F
Sbjct 158 SGVLAGSL-GTSIFDPDVFLVGASGGVYALLAAHLANVLLNYHQMRYGVIKLLHILVFVS 216
Query 242 LLFVF----------LSIGSATDV--------------VGHLAGMAAGLSFGLVYNSSNP 277
F F L +GS+++ V HLAG AGL+ GL+ S
Sbjct 217 FDFGFAIYARYAGDELQLGSSSEFLAIDQAETAGAVSYVAHLAGAIAGLTIGLLVLKSFE 276
Query 278 Q 278
Q
Sbjct 277 Q 277
> 7292669
Length=219
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/153 (28%), Positives = 82/153 (53%), Gaps = 16/153 (10%)
Query 129 IPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLG 188
I + +R + WR + +F+H+ I+HL +N+ +QI +G ++E + R+ + YL L G
Sbjct 25 IYNPYKRYEGWRFVSYMFVHVGIMHLMMNLIIQIFLGIALELVHHWWRVGLVYLAGVLAG 84
Query 189 NLVSAATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLS 248
++ ++ T G+S ++ LI +A +++ + +E ++QL+ F L+F F
Sbjct 85 SMGTSLT-SPRIFLAGASGGVYALITAHIATIIMNYSEME---YAIVQLLAF-LVFCFTD 139
Query 249 IGSA-----TD------VVGHLAGMAAGLSFGL 270
+G++ TD V HL+G AGL G+
Sbjct 140 LGTSVYRHLTDQHDQIGYVAHLSGAVAGLLVGI 172
> HsM11967983
Length=855
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 66/146 (45%), Gaps = 19/146 (13%)
Query 136 GQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAAT 195
Q +RL +FLH ILH +++ Q+ + +E+ G RI + YL+ + GNL S A
Sbjct 651 DQFYRLWLSLFLHAGILHCLVSICFQMTVLRDLEKLAGWHRIAIIYLLSGVTGNLAS-AI 709
Query 196 FFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRP---------VVLQLVLFGLLFVF 246
F +VG + + FG++ EL W + RP VVL L FGLL
Sbjct 710 FLPYRAEVGPAGSQFGILACLFVELFQSWQILA--RPWRAFFKLLAVVLFLFTFGLL--- 764
Query 247 LSIGSATDVVGHLAGMAAGLSFGLVY 272
D H++G +GL +
Sbjct 765 ----PWIDNFAHISGFISGLFLSFAF 786
> Hs21359943
Length=855
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 66/146 (45%), Gaps = 19/146 (13%)
Query 136 GQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAAT 195
Q +RL +FLH ILH +++ Q+ + +E+ G RI + YL+ + GNL S A
Sbjct 651 DQFYRLWLSLFLHAGILHCLVSICFQMTVLRDLEKLAGWHRIAIIYLLSGVTGNLAS-AI 709
Query 196 FFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRP---------VVLQLVLFGLLFVF 246
F +VG + + FG++ EL W + RP VVL L FGLL
Sbjct 710 FLPYRAEVGPAGSQFGILACLFVELFQSWQILA--RPWRAFFKLLAVVLFLFTFGLL--- 764
Query 247 LSIGSATDVVGHLAGMAAGLSFGLVY 272
D H++G +GL +
Sbjct 765 ----PWIDNFAHISGFISGLFLSFAF 786
> 7292083
Length=355
Score = 56.6 bits (135), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 68/150 (45%), Gaps = 13/150 (8%)
Query 134 RRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSA 193
RR Q+WR +FLH N HL N+ +Q+ G +E +GT RI V Y+ G+L
Sbjct 146 RRLQVWRFFSYMFLHANWFHLGFNIVIQLFFGIPLEVMHGTARIGVIYMAGVFAGSL-GT 204
Query 194 ATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLF------GLLFVFL 247
+ VG+S ++ L+ LA + + + ++ + +V+F L+
Sbjct 205 SVVDSEVFLVGASGGVYALLAAHLANITLNYAHMKSASTQLGSVVIFVSCDLGYALYTQY 264
Query 248 SIGSA------TDVVGHLAGMAAGLSFGLV 271
GSA + HL G AGL+ G +
Sbjct 265 FDGSAFAKGPQVSYIAHLTGALAGLTIGFL 294
> Hs13375799
Length=619
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 44/169 (26%), Positives = 75/169 (44%), Gaps = 9/169 (5%)
Query 137 QIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAATF 196
Q +RL +FLH ++H +++ Q+ I +E+ G RI + +++ + GNL S A F
Sbjct 416 QFYRLWLSLFLHAGVVHCLVSVVFQMTILRDLEKLAGWHRIAIIFILSGITGNLAS-AIF 474
Query 197 FCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSATDVV 256
+VG + + FGL+ EL W +E L L L + D +
Sbjct 475 LPYRAEVGPAGSQFGLLACLFVELFQSWPLLERPWKAFLNLSAIVLFLFICGLLPWIDNI 534
Query 257 GHLAGMAAGLSFGLVYNSSNPQGSPHVDVYTRISYAFNAVIVGVSVVAL 305
H+ G +GL + P++ T Y A+I+ VS++A
Sbjct 535 AHIFGFLSGLLLAFAF-------LPYITFGTSDKYRKRALIL-VSLLAF 575
> Hs4506525_2
Length=302
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 42/151 (27%), Positives = 68/151 (45%), Gaps = 18/151 (11%)
Query 135 RGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAA 194
R + WR + +F+H+ + L N +Q+ IG +E +G RI + YL L G+L +
Sbjct 106 RARAWRFLTYMFMHVGLEQLGFNALLQLMIGVPLEMVHGLLRISLLYLAGVLAGSLTVSI 165
Query 195 TFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSAT- 253
T A VG S ++ L LA +++ W + R P L ++ L+ + +G A
Sbjct 166 TDM-RAPVVGGSGGVYALCSAHLANVVMNWAGM--RCPYKLLRMVLALVCMSSEVGRAVW 222
Query 254 --------------DVVGHLAGMAAGLSFGL 270
+ HLAG G+S GL
Sbjct 223 LRFSPPLPASGPQPSFMAHLAGAVVGVSMGL 253
> At5g38510
Length=434
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/152 (28%), Positives = 70/152 (46%), Gaps = 12/152 (7%)
Query 125 FGASIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVC 184
+GA I I G+ WRL+ P+FLH I H+ ++ + + G + YG C+ Y++
Sbjct 218 YGAKINDLILAGEWWRLVTPMFLHSGIPHVALSSWALLTFGPKVCRDYGLFTFCLIYILG 277
Query 185 ALLGNLVSAATFFCNAL-KVGSSTAIFGLIGIDLAEL-----LVIWHSIEDRRPVVLQLV 238
+ GN +S F A VG + F LIG L + ++ + ED + +
Sbjct 278 GVSGNFMS---FLHTADPTVGGTGPAFALIGAWLVDQNQNKEMIKSNEYEDLFQKAIIMT 334
Query 239 LFGLLFVFLSIGSATDVVGHLAGMAAGLSFGL 270
FGL+ LS D +L + AG+ +G
Sbjct 335 GFGLI---LSHFGPIDDWTNLGALIAGIVYGF 363
> 7297676_2
Length=689
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 64/132 (48%), Gaps = 5/132 (3%)
Query 137 QIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAATF 196
Q++RL+ + +H ILHL I + Q A +E GT R + Y++ GNL S A
Sbjct 352 QLYRLLTSLCMHAGILHLAITLIFQHLFLADLERLIGTVRTAIVYIMSGFAGNLTS-AIL 410
Query 197 FCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGS---AT 253
+ +VG S ++ G++ L LLV H +P + L L V + IG+
Sbjct 411 VPHRPEVGPSASLSGVVA-SLIALLVWMHWKYLHKPHIALFKLLLLCSVLVGIGTLPYQL 469
Query 254 DVVGHLAGMAAG 265
+ +G LAG+ G
Sbjct 470 NFLGLLAGVICG 481
> CE24148
Length=851
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 40/136 (29%), Positives = 62/136 (45%), Gaps = 9/136 (6%)
Query 136 GQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAAT 195
QI+RL +F+H ++HL ++M Q+ A E G++R+ + Y + GNL S A
Sbjct 573 NQIYRLFTSLFIHAGVIHLALSMAFQMYFMAYQENLIGSKRMAILYFASGISGNLAS-AI 631
Query 196 FFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQL-----VLFGLLFVFLSIG 250
F VG S+A G+ + EL WH P L+ ++ LL + + +
Sbjct 632 FVPYYPTVGPSSAQCGVFSSVVVEL---WHFRHLLDPFELKFQSIAHLIVTLLVLCIGLI 688
Query 251 SATDVVGHLAGMAAGL 266
D HL G GL
Sbjct 689 PWIDNWSHLFGTIFGL 704
> CE29062
Length=727
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 61/131 (46%), Gaps = 1/131 (0%)
Query 136 GQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAAT 195
Q +RL +F+H ++HL +++ Q + +E ++R+ + Y + GNL S A
Sbjct 450 NQFYRLFTSLFVHAGVIHLALSLLFQYYVMKDLENLIASKRMAILYFASGIGGNLAS-AI 508
Query 196 FFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSATDV 255
F VG S+A G++ + E I++ + ++Q ++ LL + + D
Sbjct 509 FVPYNPAVGPSSAQCGILAAVIVECCDNRRIIKEFKWALVQHLIVTLLVLCIGFIPWVDN 568
Query 256 VGHLAGMAAGL 266
HL G GL
Sbjct 569 WAHLFGTIFGL 579
> Hs17445238
Length=619
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 46/93 (49%), Gaps = 1/93 (1%)
Query 136 GQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAAT 195
Q +RL +FLH ILH +++ Q+ I +E+ G I + YL+ + GNL S A
Sbjct 427 DQFYRLWLSLFLHTRILHYLVSICFQMTIPRDLEKLAGWHHIAIIYLLSGVTGNLAS-AI 485
Query 196 FFCNALKVGSSTAIFGLIGIDLAELLVIWHSIE 228
F +V + + FG++ EL W +E
Sbjct 486 FLLYRAEVVPAGSQFGILACLFMELFQSWQILE 518
> At3g58460
Length=411
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 40/154 (25%), Positives = 73/154 (47%), Gaps = 14/154 (9%)
Query 129 IPSAI-RRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCA-- 185
+PSAI R Q++R + H ++LH+ NM + +G+ +E G+ R+ ++ A
Sbjct 54 LPSAIISRFQVYRFYTAIIFHGSLLHVLFNMMALVPMGSELERIMGSVRLLYLTVLLATT 113
Query 186 ------LLGNLVSAATFF-----CNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVV 234
L+ +L F+ N +G S +F +I I+ + V S+ V
Sbjct 114 NAVLHLLIASLAGYNPFYQYDHLMNECAIGFSGILFSMIVIETSLSGVTSRSVFGLFNVP 173
Query 235 LQLVLFGLLFVFLSIGSATDVVGHLAGMAAGLSF 268
+L + LL VF + + ++GHL G+ +G S+
Sbjct 174 AKLYPWILLIVFQLLMTNVSLLGHLCGILSGFSY 207
> CE00492
Length=397
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 42/175 (24%), Positives = 77/175 (44%), Gaps = 25/175 (14%)
Query 117 PSISSLITFGASIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRR 176
P++S LI +P ++WRL +++ I H+ N+ +Q+ IG +E + R
Sbjct 116 PTMSPLIVSQYHLP------ELWRLFTYCLINVGIFHIIFNILIQLAIGVPLELVH-RWR 168
Query 177 ICVAYLVCALLGNLVSAA----TFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRP 232
I + Y + L G+++S A F C G + F LI + + + +E+
Sbjct 169 IYILYFMGVLFGSILSLALDPTVFLC-----GGAAGSFSLIASHITTIATNFKEMENATC 223
Query 233 VVLQLVLFGLLFVFLSIG--------SATDVVGHLAGMAAGLSFGLV-YNSSNPQ 278
+ L++F L L++ + GHL G+ AG+ F + + S P
Sbjct 224 RLPILIVFAALDYVLAVYQRFFAPRIDKVSMYGHLGGLVAGILFTFILFRGSKPS 278
> Hs8923409
Length=292
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 70/153 (45%), Gaps = 14/153 (9%)
Query 129 IPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLG 188
I S +R + WR I + +H + H+ N+ MQ+ +G +E + R+ + YL + G
Sbjct 100 IYSPEKREEAWRFISYMLVHAGVQHILGNLCMQLVLGIPLEMVHKGLRVGLVYLAGVIAG 159
Query 189 NLVSAATFFCNALKVGSSTAIFGLIGIDLAELLV----------IWHSIEDRRPVVLQL- 237
+L S + F VG+S ++ L+G +LV I+ + +VL +
Sbjct 160 SLAS-SIFDPLRYLVGASGGVYALMGGYFMNVLVNFQEMIPAFGIFRLLIIILIIVLDMG 218
Query 238 -VLFGLLFVFLSIGSATDVVGHLAGMAAGLSFG 269
L+ FV GS H+AG AG+S G
Sbjct 219 FALYRRFFV-PEDGSPVSFAAHIAGGFAGMSIG 250
> Hs10190734_1
Length=292
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 44/197 (22%), Positives = 87/197 (44%), Gaps = 17/197 (8%)
Query 79 CLLPKFNAKHCVFILAVIDLIVYIATVIYGQSPDNPTSPSISSLITFGASIPSAIRRGQI 138
CL P+ F A++ L+V + Q P P+ ++ S A+R Q+
Sbjct 57 CLCPE--VPSATFFTALLSLLVSGPRLFLLQQPLAPSGLTLKS---------EALRNWQV 105
Query 139 WRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAATFFC 198
+RL+ +F++ N + L + R + E GT R C ++ A+ ++ +
Sbjct 106 YRLVTYIFVYENPISLLCGAIIIWRFAGNFERTVGTVRHCFFTVIFAIFSAIIFLSFEAV 165
Query 199 NAL-KVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSA----- 252
++L K+G G + A L V RR +V +V+ +L +L +G++
Sbjct 166 SSLSKLGEVEDARGFTPVAFAMLGVTTVRSRMRRALVFGMVVPSVLVPWLLLGASWLIPQ 225
Query 253 TDVVGHLAGMAAGLSFG 269
T + ++ G++ GL++
Sbjct 226 TSFLSNVCGLSIGLAYA 242
> 7292077
Length=235
Score = 43.1 bits (100), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 45/154 (29%), Positives = 71/154 (46%), Gaps = 21/154 (13%)
Query 134 RRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSA 193
+R Q+WR + LH + LHL N+ Q+ G +E +G+ R V Y+ L G+L
Sbjct 25 QRLQLWRFLSYALLHASWLHLGYNVLTQLLFGVPLELVHGSLRTGVIYMAGVLAGSL-GT 83
Query 194 ATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLFGLLFVFLSIGSA- 252
+ VG+S ++ L+ LA LL+ + R V+QL+ +LFVF +G A
Sbjct 84 SVVDSEVFLVGASGGVYALLAAQLASLLL---NFGQMRHGVIQLMAV-ILFVFCDLGYAL 139
Query 253 ---------------TDVVGHLAGMAAGLSFGLV 271
+ H+ G AG+S GL+
Sbjct 140 YSRELAMHQLQTRPSVSYIAHMTGALAGISVGLL 173
> 7297927
Length=263
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 50/94 (53%), Gaps = 1/94 (1%)
Query 137 QIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAATF 196
+ WRL+ + LH + HL +N+ Q IG +E + G R+ V Y+V + G+L + A
Sbjct 106 EYWRLLTYMLLHSDYWHLSLNICFQCFIGICLEVEQGHWRLAVVYMVGGVAGSLAN-AWL 164
Query 197 FCNALKVGSSTAIFGLIGIDLAELLVIWHSIEDR 230
+ +G+S ++ ++G + L++ + + R
Sbjct 165 QPHLHLMGASAGVYAMLGSHVPHLVLNFSQLSHR 198
> 7303544
Length=351
Score = 35.4 bits (80), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 44/98 (44%), Gaps = 5/98 (5%)
Query 120 SSLITFGASIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICV 179
S++IT+ S P+A + W + F H + +HLF NM++ + G +
Sbjct 169 STMITYFTSNPAA--KVVCWPMFLSTFSHYSAMHLFANMYVMHSFANAAAVSLGKEQFLA 226
Query 180 AYLVCALLGNLVS---AATFFCNALKVGSSTAIFGLIG 214
YL + +L+S A + +G+S AI L+
Sbjct 227 VYLSAGVFSSLMSVLYKAATSQAGMSLGASGAIMTLLA 264
> At3g59520
Length=269
Score = 34.7 bits (78), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 48/181 (26%), Positives = 73/181 (40%), Gaps = 45/181 (24%)
Query 126 GASIPSAIRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAY---- 181
G S +AI G WR+I H+++LHL NM +G + E+ G + AY
Sbjct 40 GLSYETAIE-GHYWRMITSALSHISVLHLVFNMSALWSLG--VVEQLGHVGLGTAYYLHY 96
Query 182 ----------LVCALLGNLVS--AATFFCNALKVGSSTAIFGLIGIDLAELLVIWHSIED 229
LV + L++ +F VG S +FG W +I
Sbjct 97 TLVLVVFSGVLVIGIYHLLIARFKIDYFRRVTAVGYSCVVFG------------WMTILS 144
Query 230 RRPVVLQLVLFGLL-----------FVFLS-IGSATDVVGHLAGMAAG--LSFGLVYNSS 275
+ +L LFGLL +F S I +GHL+G+ G +S+GL+ +
Sbjct 145 VKQPSSKLNLFGLLSLPISFAPFESLIFTSIIVPQASFLGHLSGILVGYAISWGLIGGMN 204
Query 276 N 276
N
Sbjct 205 N 205
> SPBP4H10.10
Length=392
Score = 33.5 bits (75), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 28/60 (46%), Gaps = 0/60 (0%)
Query 133 IRRGQIWRLICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVS 192
+ G+ W L+ +F H N+ HL +N + + K+G + YL + GN V+
Sbjct 164 LYEGRWWTLVVSIFSHQNLAHLLVNCVAIYSFLSIVVYKFGVWKALSVYLGAGVFGNYVA 223
> CE16121
Length=1049
Score = 30.8 bits (68), Expect = 3.6, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 29/62 (46%), Gaps = 4/62 (6%)
Query 92 ILAVIDLIVYIATVIYGQSPDNPTSPSISSLITFGASIPSAIRRGQIWRLICPVFLHLNI 151
IL ID+ + G+S D PT PS SS+ S R+G W L+ V+ +
Sbjct 147 ILTKIDISKDLKGERLGESQDEPTDPSTSSI----TSDEQLRRKGLSWLLMSDVYEEAFV 202
Query 152 LH 153
LH
Sbjct 203 LH 204
> At5g25640
Length=102
Score = 30.4 bits (67), Expect = 4.8, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 165 GASIEEKYGTRRICVAYLVCALLGNLVSAATFFCNALKVGSSTAIFGLIGIDL 217
G +EE+ G + ++YL + N+VS T N+ G+S A+FGL I +
Sbjct 9 GKIVEEEKGNFGLWLSYLFTGVGANMVSWFTLPPNSFFSGASGAVFGLFVISV 61
> YGR101w
Length=346
Score = 30.0 bits (66), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 33/79 (41%), Gaps = 5/79 (6%)
Query 141 LICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVS-----AAT 195
+I F H HL +NM G S+ G Y+ A+ G+L S A
Sbjct 186 IIGSAFSHQEFWHLGMNMLALWSFGTSLATMLGASNFFSLYMNSAIAGSLFSLWYPKLAR 245
Query 196 FFCNALKVGSSTAIFGLIG 214
+G+S A+FG++G
Sbjct 246 LAIVGPSLGASGALFGVLG 264
> Hs20127652_2
Length=208
Score = 29.6 bits (65), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 21/100 (21%), Positives = 40/100 (40%), Gaps = 4/100 (4%)
Query 141 LICPVFLHLNILHLFINMFMQIRIGASIEEKYGTRRICVAYLVCALLGNLVSAATFFCNA 200
++ F H ++ H+ NM++ +SI G + YL ++ N VS +
Sbjct 38 MLLSTFSHFSLFHMAANMYVLWSFSSSIVNILGQEQFMAVYLSAGVISNFVS----YVGK 93
Query 201 LKVGSSTAIFGLIGIDLAELLVIWHSIEDRRPVVLQLVLF 240
+ G G G + L + I + R ++ L +F
Sbjct 94 VATGRYGPSLGASGAIMTVLAAVCTKIPEGRLAIIFLPMF 133
> Hs4503359
Length=412
Score = 29.6 bits (65), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 3 RQQHRKAAPTGIATTTILILSQRQTNQGWQQQQRREQGPAGLM 45
R H P G+A ++ Q + W++Q R++ PAGL
Sbjct 340 RPDHIYDKPEGVAALSLYDSPQEPRGEAWRRQATRDRDPAGLQ 382
Lambda K H
0.328 0.140 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7237550830
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40